Enzyme's 'editing' preferences have implications for infertility and cancer

To "turn off" particular regions of genes or protect them from damage, DNA strands can wrap around small proteins, called histones, keeping out all but the most specialized molecular machinery. Now, new research shows how an enzyme called KDM4B "reads" one and "erases" another so-called epigenetic mark on a single histone protein during the generation of sex cells in mice. The researchers say the finding may one day shed light on some cases of infertility and cancer.
A summary of the work, a collaborative study among researchers at the Johns Hopkins University School of Medicine, Rice University and the University of Wisconsin-Madison, was published online in the journal Nature Communications on Nov. 14.
"Our research gives us insight into how cells use this epigenetic machinery to shut off and protect parts of the genome during reproduction," says Sean Taverna, Ph.D., associate professor of pharmacology and molecular sciences at the Johns Hopkins University School of Medicine. "Because KDM4B and its histone target are prevalent in mouse testes, and the absence of that histone target inhibits reproduction in a single-celled organism, we think this interaction could play a role in human infertility or other diseases."
At the heart of their new discovery, says Taverna, is the fact that genes housed in a cell's nucleus are not the whole story when it comes to the proteins they ultimately make. If genes are the sentences of a book, the story can be altered dramatically depending on which sentences are read and skipped over, and in what order. Histone proteins, Taverna explains, act like removable tape sealing closed particular pages of the book. When DNA is wrapped around them, genetic regions are "silenced" and go unread by other proteins—a classic epigenetic control mechanism, since it changes how the genome is read without changing its sentences.
Histones are frequently controlled through small chemical "tags," especially methyl groups, which can be attached to and detached from specific spots on histone proteins. To advance its understanding of this process, Taverna's lab had previously turned to a one-celled organism, Tetrahymena thermophila, with an unusual trait: It has not one but two nuclei. The DNA in one nucleus is active and used for making proteins, while the DNA in the other is "silent" except during mating. That research revealed a new site for methyl groups on one histone. That spot, a lysine known as H3K23, has little to no methyl tags in active Tetrahymena nuclei but is loaded up with three methyl groups (H3K23me3) in "mating nuclei" undergoing early meiosis, the special version of cell division that creates sex cells.
In those experiments on Tetrahymena, the team learned that H3K23me3 appears in regions of the genome that should never be cut by enzymes during meiosis. DNA cuts normally occur during meiosis to allow for the mixing of genetic material and the proper sorting out of chromosomes in dividing cells, so H3K23me3 could protect nearby regions of the genome by "taping them up" so they can't be accessed.
In a hunt for what controls that part of the process, researchers at the University of Wisconsin-Madison led by John Denu, Ph.D., developed a way to find out which proteins bind to which histone tags. Specifically, they identified a family of three similar enzymes called lysine demethylases, or KDM4s, which have been implicated in the development of some cancers in humans. KDM4A binds H3K23me3 plus two other histone tags and KDM4C binds another histone tag, but KDM4B binds only H3K23me3.
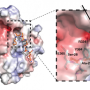
The researchers wondered what feature enables the three KDM4s to be so selective in their binding, so colleagues at Rice University, led by George Phillips, Ph.D., picked up the research baton at that point to understand their selectivity by solving their 3-D structures.
While KDM4B's structure proved elusive, the scientists were able to figure out the structure of KDM4A bound to H3K23me3. By comparing the differences between the KDM4s and by tweaking three of KDM4A's amino acid building blocks to make it act like KDM4B, the scientists report they were able to learn the basis for the binding preferences of all three KDM4s.
"Now that we know the 'rules' that determine how KDM4s bind, we can imagine ways to intervene to change their interactions with histone tags," says Taverna. "That in turn means we can test what happens when we enhance their activity or turn it down, and better understand their roles in disease."
To that end, biochemical tests revealed that after KDM4B "reads" H3K23me3, it "erases" the methyl groups from a nearby lysine, H3K36me3. The researchers say that more work is needed to understand the significance of this rare combination of activities, but Taverna speculates that the "demethylation"—that is, removing the epigenetic marks—of H3K36 helps to "tape up," silence and protect nearby DNA.
"We know from our work in Tetrahymena that H3K23me3 is most often found in regions of the genome that should never suffer DNA damage because that would harm reproduction," he says. "During meiosis, when H3K23me3 is most prevalent, DNA is prone to damage, which explains why Tetrahymena lacking H3K23me3 struggle to reproduce."
To see if H3K23me3 plays a role in mammalian reproduction, Taverna's group, led by postdoctoral fellow Kimberly Stephens, Ph.D., looked for its prevalence in rodents and found it and KDM4B to be increased in immature sperm undergoing early meiosis. (Since early meiosis occurs in females as they develop in the womb, the team was unable to examine immature eggs.)
"While our experiments offer no proof that defects in the KDM4B-H3K23me3 interaction cause infertility in mammals, all of the signs point in that direction," says Taverna, "and we definitely plan to study it further."
More information: Zhangli Su et al, Reader domain specificity and lysine demethylase-4 family function, Nature Communications (2016). DOI: 10.1038/ncomms13387