New insights into mechanisms of breast cancer development and resistance to therapy
Why does breast cancer develop and how come certain patients are resistant to established therapies? Researchers from the University of Basel have gained new insights into the molecular processes in breast tissue. They identified the tumor suppressor LATS as a key player in the development and treatment of breast cancer. The journal Nature has published the results today.
All breast cancers are not created equal. In up to 70 percent of all breast cancers, the tumor has receptors for the hormone estrogen. Today, these estrogen-receptor-positive cancers can be treated relatively well. Because these tumors need estrogen for their growth, the receptor is the target of a number of drugs that interfere with estrogen expression, bind to the receptor or speed up its degeneration.
However, around a third of all patients does not react to therapy or develops resistance. So far it has not been possible to accurately predict who will respond to this therapy because the underlying molecular mechanisms are not yet understood entirely.
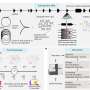
In a comprehensive molecular study, a group of scientists led by Prof. Mohamed Bentires-Alj from the Department of Biomedicine at the University and the University Hospital of Basel has now identified an important player in this process named LATS. They were able to show how this enzyme, in cooperation with other proteins, influences the development and treatment of breast cancer.
Tumor suppressor LATS decides cell fate
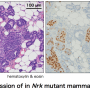
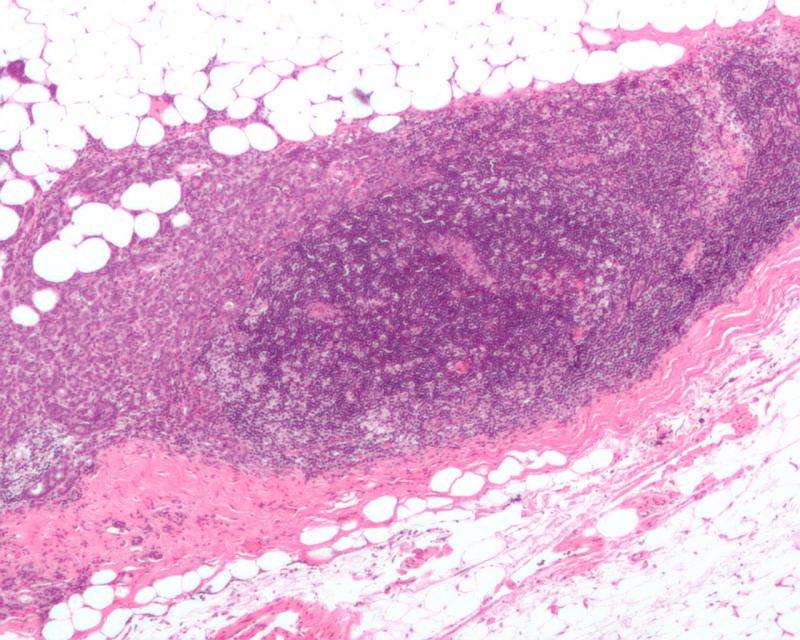
The researchers focused on cancer-inhibiting genes that prevent normal cells from becoming cancerous. In particular, they studied the tumor suppressors LATS1 and LATS2. Once LATS is deleted, the processes in the breast tissue change.
Without LATS, the number of so-called luminal precursor cells in the epithelial tissue of breast glands increases. These are the cells of origin of most types of breast cancer in humans. "LATS balances cell fate in the breast tissue. In its absence the equilibrium shifts and more cells that can give rise to tumors develop", explains Bentires-Alj.
Resistance to degradation
In healthy breast tissue, LATS brings together the estrogen receptor alpha with the protein degradation machinery. Without LATS the receptor can no longer be properly degraded, which has consequences for cancer therapy. "We were able to show that cancer cells without LATS no longer respond to Fluvestrant, an estrogen-receptor antagonist that promotes its degradation. They were resistant", says Bentires-Alj.
The removal of LATS also stabilized the proteins YAP and TAZ, which are upregulated in many cancers and boost cell proliferation. "Thanks to our newly gained insights into the molecular processes in healthy breast tissue, we now also better understand how cells of origin of cancer expand and why certain tumors are resistant to therapy", summarizes the Basel scientists Bentires-Alj.
More information: Adrian Britschgi et al, The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERα, Nature (2017). DOI: 10.1038/nature20829