Sumo protein explored as likely source for some congenital heart defects

Small ubiquitin-like modifier (SUMO) proteins are small peptides that get added on to other proteins to regulate their activity. While SUMO has many regulatory roles in cells, it is especially important for controlling gene expression during early development. Just a few years ago this connection between SUMO and gene regulation was relatively unknown, but now, Notre Dame researchers are exploring how a disruption to the SUMO protein's ability to regulate embryo development may be linked to congenital heart defects.
Paul Huber, professor of chemistry and biochemistry, and Norman Dovichi, the Grace-Rupley Professor of Chemistry and Biochemistry, are working together to understand the role of all proteins in embryo development using Xenopus laevis or the African claw frog. This species is known for having a similar gene structure to that of the human genome, meaning that findings related to this species have the potential to provide a deep understanding about human diseases.
When discussing their research, Dovichi said, "In 2014, Huber and I completed a study using Xenopus laevis embryos to understand how more than 4,000 proteins fluctuate during the different stages of development. We found that certain proteins spike or lower during specific stages. For example, a number of proteins that are used during the creation of cardiovascular tissue rose during stage 13, when organs develop."
In these early experiments, Huber found that when SUMO activity was repressed, many of the embryos had two predominate phenotypes, one of which was heart defects. Then, new studies began linking mutations in SUMO protein to heart failure as well as congenital heart defects. This is when the Notre Dame researchers began to develop the next steps for their research.
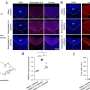
"At the time, there was not a lot of information about the role of the SUMO protein, but our theory was that it was critical for proper development of the heart," said Huber. "To study the protein's specific impact, we inhibited SUMO activity in the developing cardiovascular tissue. This will allow us to compare the proteome – or all of the proteins that are expressed by a cell – of the defective hearts with their normal counterparts."
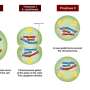
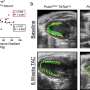
To support the research, Olivia Cox, a Notre Dame graduate student, collaborated with Daniel Weeks, professor of biochemistry and pediatrics at the University of Iowa, and identified three predominate heart defects in the SUMO-deficient hearts: septal defects – commonly thought of as holes in the heart – abnormal turning of the outflow tract, and noncompaction cardiomyopathy.
The goal of this research is to use the proteome comparisons to specifically identify which proteins are responsible for congenital heart defects. Additionally, Huber, affiliated with the Harper Cancer Research Institute (HCRI) and the Center for Stem Cells and Regenerative Medicine, and Dovichi, affiliated with Advanced Diagnostics and Therapeutics as well as the HCRI, plan to continue exploring the SUMO protein's significance in other areas of embryo development. This research could help explain why other development defects arise, and eventually lead to a solution for increasing SUMO protein expression when a mutation occurs.
More information: Liangliang Sun et al. Quantitative proteomics of Xenopus laevis embryos: expression kinetics of nearly 4000 proteins during early development, Scientific Reports (2014). DOI: 10.1038/srep04365