Research on crucial cutting enzyme maps sites of DNA damage in leukemias and other cancers

Researchers studying a DNA-cutting enzyme with a crucial role in regulating the structure of genes have discovered a broad role for its cutting activity in driving abnormal genetic rearrangements called translocations that cause cancer, including leukemias and solid tumors.
The enzyme, type II topoisomerase, called TOP2, snips the DNA double strand to allow fundamental cellular activities to occur: transcription (converting DNA information into RNA) and replication (copying a DNA double helix into two daughter molecules). TOP2 also rejoins the broken DNA strands that it cuts, but if this process goes wrong, mismatched ends of DNA can give rise to a translocation. The translocation can produce a fusion protein leads to abnormal white blood cells, the hallmark of leukemia.
A physician-researcher and a biologist specializing in RNA have teamed up to create a novel DNA sequencing tool that identifies specific sites of TOP2 cutting (cleavage) along the entire genome of human leukemia cells. "This tool opens new possibilities to better understand and eventually manipulate TOP2 cutting to prevent the rearrangements that give rise to leukemias," said study co-leader Carolyn A. Felix, MD, a pediatric oncologist at Children's Hospital of Philadelphia. Felix's research focuses on infant leukemia as well as treatment-related or secondary leukemia that may occur as a side effect of TOP2-related chemotherapy drugs.
Felix's team has collaborated for the past several years with a team led by biologist Brian D. Gregory, PhD, of the University of Pennsylvania, who uses genome-wide sequencing to catalog the sites on DNA and RNA molecules that are bound by proteins. "We designed a way to pull down the DNA bound to TOP2, then to break that bond so that only the DNA undergoing cleavage is free to be sequenced at single base-pair precision," he said. "This enabled us to map, for the first time, topoisomerase cleavage on a genome-wide scale."
The scientists reported their findings online April 6, 2017 in the journal Genome Research.
For some time, oncologists have used drugs called TOP2 poisons that harness TOP2's DNA-cutting activity to damage DNA and kill cancer cells. But TOP2 poisons are a double-edged sword, because intervening in the cutting and rejoining of DNA can lead to translocations, when DNA strands swap genetic material between chromosomes and rejoin incorrectly. The gene most often rearranged in these translocations in treatment-related leukemia is KMT2A.
Besides their use in chemotherapy drugs, TOP2 poisons occur in the environment and in the diet. Felix and her collaborators have previously shown that a fetus's exposure to naturally occurring TOP2 poisons in the mother's diet during pregnancy is associated with an increased risk of leukemia in infants. Similar to treatment-related leukemia caused by TOP2 poison chemotherapies, KMT2A translocation also is the hallmark abnormality in leukemia in infants. Felix previously developed a method to identify partner genes that fuse with KMT2A to form translocations. This tool pinpoints the exact position of translocation breakpoints.
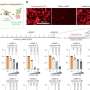
Her collaboration with Gregory's team offered the opportunity to map, with unprecedented detail, the sites of TOP2 cutting in human cells all across the genome—and to use this information to ask how TOP2 cutting is related to translocations. Their study, which was conducted in a human leukemia cell line, shows that TOP2 cutting sites along the length of genes occur in clusters, often within introns, the non-coding portion of protein-coding genes.
The researchers found evidence implicating TOP2's cutting role not just in KMT2A translocations in leukemia, but also in other cancer-driving translocations. They write that this discovery raises "the intriguing possibility that TOP2A has a previously unrecognized broader role in triggering translocations."
They also discovered TOP2 cutting in long non-coding RNAs called lincRNAs, elements in the genome that are transcribed into RNA even though they do not produce proteins. Furthermore, when the team mapped the TOP2 cutting sites in the leukemia cell line and compared these to data from the National Human Genome Research Institute's ENCODE project, they found that these sites occur in the same parts of genes as protein tags called histones that play important roles in regulating gene transcription.
"The fact that TOP2 cleavage clusters in the same genomic environment alongside histone marks and transcription regulators implicates the TOP2 enzyme as a prominent orchestrator of DNA remodeling," said Felix.
Felix added that the findings open possibilities for new clinical approaches. "The better we identify where cleavage occurs across the genome, the better we can understand how the translocations happen. We could use that knowledge to design smarter anticancer drugs to target the TOP2 enzyme that don't have such a high risk of causing translocations, or we could design drugs to protect sequences in the genome from unwanted cutting."
More information: Xiang Yu et al, Genome-wide TOP2A DNA cleavage is biased toward translocated and highly transcribed loci, Genome Research (2017). DOI: 10.1101/gr.211615.116