Different strains of same bacteria trigger widely varying immune responses
Genetic differences between different strains of the same pathogenic bacterial species appear to result in widely varying immune system responses, according to new research published in PLOS Pathogens.
Previous research has found that different people vary in their susceptibility to infection with the same species of pathogenic bacteria. Individual differences in people's immune systems may explain this variability, but differences between bacterial strains could play a role, too.
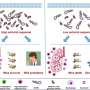
To better understand this role, Uri Sela of The Rockefeller University, New York, and colleagues studied different strains of two major species of pathogenic bacteria: Staphylococcus aureus and Streptococcus pyogenes. They tested how immune system T and B cells in donated human blood samples responded after exposure to different strains of each species.
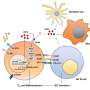
The researchers found that, in blood from a single person, different strains of each species produced widely varied responses by T and B cells of the adaptive immune system—the portion of the immune system responsible for creating "memory" of specific pathogens to protect against future infection. The same distinct responses to different strains were seen in blood samples from 10 additional donors.
Next, the research team created mutant bacteria in which they deactivated "accessory" genes that are responsible for between-strain differences, leaving the "core" genome of the species intact. They found that the mutant strains triggered a dampened T cell response, suggesting that differences in "accessory" genes were responsible for the varied responses seen for unmutated strains.
These findings suggest that differences in bacterial "accessory" genes—not just differences between people—may help explain the clinical variation generally found among patients infected with the same bacterial species.
Previous research has often described "signature" immune responses to different bacteria using only a single strain for each species. However, based on findings of the current study, the researchers propose that immune response signatures should instead be defined according to the specific strain or the species' common "core genome." Such a shift could aid development of strategies for predicting disease outcomes in patients.
"The current practice with infected patient is to only identify the bacterial species," the authors elaborate. "Our findings raise the possibility that in the future we might need to define the specific infecting strain as part of the patient evaluation and treatment."
More information: Sela U, Euler CW, Correa da Rosa J, Fischetti VA (2018) Strains of bacterial species induce a greatly varied acute adaptive immune response: The contribution of the accessory genome. PLoS Pathog 14(1): e1006726. doi.org/10.1371/journal.ppat.1006726