Technique allows live imaging of ubiquitin protein during cellular housekeeping
Homeostasis refers to the idea that cells have evolved an intricate set of systems allowing them to respond to their environment while maintaining an internal, physiologically healthy balance. These systems are especially important when cells are stressed—for example, during exposure to extreme temperatures or UV radiation from the sun—as quite a lot can go wrong if cells are unable to respond to environmental stressors.
Autophagy is a process that cells use to maintain homeostatic balance. Translating as "to devour oneself," autophagy breaks down toxic materials that build up inside of cells under duress. For example, autophagy removes proteins that fail to fold properly, which would otherwise form harmful aggregates. Autophagy is, in effect, a kind of cellular housekeeping protein.
In a study published in the journal Autophagy, researchers at Tokyo Medical and Dental University (TMDU) describe a new experimental method that may illuminate the molecular details of autophagy. Their focus was on ubiquitin, a key signaling molecule in this process.
Ubiquitin is a tiny protein that attaches itself to other proteins in the cell, like misfolded proteins, and targets them to be degraded. Ubiquitin is added in repeating units, creating a ubiquitin "tail" that the autophagy system can recognize. This stepwise process, called ubiquitination, is critical for the cell to determine which proteins need to be removed. Many complicated arrangements of ubiquitin tails can form on these proteins, though, and only some of them have been studied in detail.
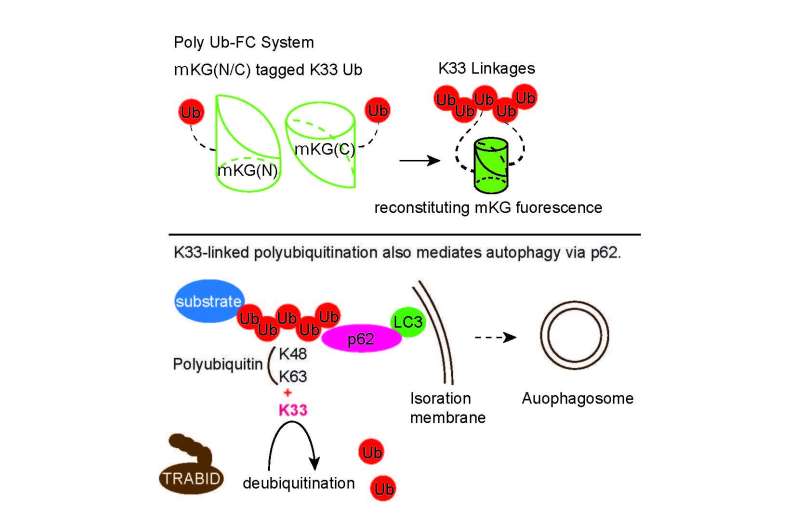
"Ubiquitin actually has eight different residues that it can potentially use when it forms a polyubiquitin chain on a target protein," lead author Yoichi Nibe explains. "We know that the particular residue used for linkage can determine which degradation pathway a protein will follow during autophagy, but so far only two of these residues have been characterized. The function of the other six is largely unexplored territory. What we needed was a system that would allow us to analyze any ubiquitin residue we choose."
To achieve this, the research team devised a system that expresses two genetically engineered ubiquitin molecules at once. The key is that each ubiquitin is fused to one-half of a fluorescent protein called Kusabira Green. When both halves of Kusabira Green are next to each other, green light is emitted—so the net result is that a cell will light up wherever the two ubiquitin molecules are attached to one another.
"Genetic tagging with a split-fluorescent protein is not in itself a new technique," corresponding author Shigeru Oshima notes. "The difference is that this system allows us to visualize ubiquitin assembly in real time, in living cells. We can tightly control the expression of ubiquitin using experimental induction, which is ideal for studying a highly dynamic process such as ubiquitination. Most importantly, though, we can manipulate individual residues of ubiquitin to force specific chains to form in the cell."
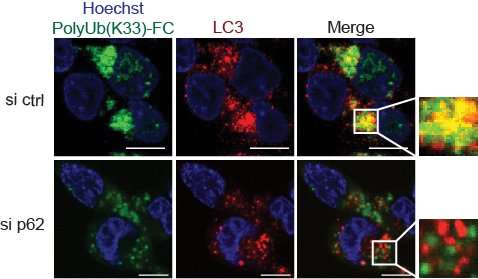
To date, only two residues involved in forming ubiquitin chains, lysine 48 and lysine 63, have been linked to autophagy. The new system allowed the team to add a third residue—lysine 33—to ubiquitin's repertoire in controlling this cellular housekeeping system. The team is hopeful that their imaging technique will eventually bear even more fruit.
"The system is very versatile," Oshima adds. "Given the complexity of ubiquitination, we expect to uncover some interesting functions in the future."
More information: Yoichi Nibe et al, Novel polyubiquitin imaging system, PolyUb-FC, reveals that K33-linked polyubiquitin is recruited by SQSTM1/p62, Autophagy (2017). DOI: 10.1080/15548627.2017.1407889