November 30, 2018 report
Researchers use optimized single-cell multi-omics sequencing to better understand colon cancer tumor heterogeneity
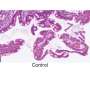
A team of researchers affiliated with several institutions in China has found that using optimized single-cell multi-omics sequencing better reveals colon cancer tumor heterogeneity. In their paper published in the journal Science, the group describes their unique approach to understanding colorectal cancer progression.
The researchers note that most genetic studies of colorectal cancer progression involve looking at gene expression. They suggest that more study is needed to learn how colorectal tumors metastasize. To that end, they have developed a sequencing method that allows for analyzing copy number variants, methylation and gene expression simultaneously in individual cells—the method combines single-cell sequencing data with information from chromosome conformation, epigenetic data and other characteristics of tumor cells.
This work is the next step in a long-term effort to truly understand the mechanics of metastases, particularly in colorectal tumors. Two years ago, the team published a report on their work involving a single-cell triple omics sequencing technique they had developed called sc Trioseq by which they gathered information from gene expression, methylation at CpG sites and copy number alterations from 25 cells obtained from cancer patients.
In the next stage, the researchers raised the number of cells to 1,900 and improved the efficiency of the detection method. The study consisted of collecting cell samples from 12 patients, 10 of whom provided both primary and metastatic data and analyzing them. Using cell data from both sources allowed the researchers to isolate and identify genetic lineages that had developed from mutations for each patient. They used methylation data and copy number information to identify those lineages, allowing them to track the evolutionary changes they went through as they moved from primary tumor cells to metastatic cells.
The team reports that methylation was consistent among the cells within the same genetic lineage but differed when compared to other lineages—and it was also different from non-tumorous cells just next to the tumor. They also found that six chromosomes showed greater levels of demethylation than did others, three of which had recurrent chromosomal changes. They conclude by suggesting that single cell multi-omics sequencing offers the best opportunity for learning more about tumor progression and the spread of cancer.
More information: Shuhui Bian et al. Single-cell multiomics sequencing and analyses of human colorectal cancer, Science (2018). DOI: 10.1126/science.aao3791
© 2018 Science X Network