Columbia professor uses data science to create personalized cancer therapies
Columbia's Andrea Califano has developed a highly-predictive computer platform that can analyze all tumor types and predict which drug or drugs will be most effective in treating them.
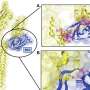
Califano's lab uncovered so-called master regulators in these tumor cells by using advanced data science techniques. They first used a branch of mathematics called "information theory" to identify the genes regulated by every protein in GEP-NET cells, thus establishing a gene regulatory network for this type of tumors. Then they used the VIPER algorithm—a method akin to ray-tracing, i.e., tracing each light ray back from an object's surface to the source that illuminates it. The technique is used by the film industry to render realistic three-dimensional objects. VIPER allowed the team to navigate backward through the gene regulatory networks to identify the specific proteins that physically control the molecular signature of a patient's tumor. These are the critical proteins dubbed master regulators. The Califano Lab and Modlin collaborated on the research with Columbia's Department of Pathology and Cell Biology, Columbia's Department of Urology, the Broad Institute of Harvard and MIT as well as 17 other research institutions.
"This research represents a first proof of concept for what may become a valuable new tool to deliver an effective and systematic precision-medicine approach to cancer patients," says Califano, the Clyde and Helen Wu Professor of Chemical and Systems Biology and chair of the Department of Systems Biology at Columbia University Irving Medical Center (CUIMC). He is also a member of the Data Science Institute's Health Analytics Center at Columbia.
Califano is also part of a research team from CUIMC that recently received a 2018 Prostate Cancer Foundation (PCF) Challenge Award from the Prostate Cancer Foundation to advance prostate cancer research by studying master regulators that underlay tumor-micro environment interactions in metastatic prostate cancer. The two other principal investigators are Michael Shen and Charles Drake. The three researchers will combine their expertise in systems biology, basic science, immunology, pathology and medicine to develop precision-medicine treatments that target the prostate tumor micro environment.
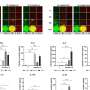
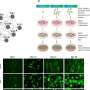
Califano's lab additionally developed a computational framework, called OncoTreat, that can predict the drug or drugs that inhibit the activity of the master regulators. He says that oftentimes the same cancer-driving proteins—the master regulators—that his team discovered in one cancer type are active in patients with different kinds of cancer, an assessment based on the team's analysis of tumor tissues from more than twenty thousand patients. The tumor tissues were part of the Cancer Genome Atlas repository assembled by the National Cancer Institute. The team has also conducted experiments on mice to determine which of 120 FDA-approved drugs and 340 experimental compounds prove effective against cancer cells by targeting their master-regulator proteins. These experiments also allowed the team to develop the computer-based OncoTreat algorithm, which then recommends treatments for cancer patients based on the specific set of master regulator proteins that drive their tumors. The OncoTreat algorithms as well as VIPER have been validated in a series of papers published by Califano's lab.
Califano's research is garnering national recognition. He was recently elected to the National Academy of Medicine for his work on master regulators. And based on his research findings, the FDA recently approved a clinical trial at Columbia for patients with GEP-NET. During the initial phase of the trial, dozens of patients with this type of neuroendocrine tumors will have their protein levels assessed by Califano's team. His lab will also test a drug, entinostat, that was predicted to accurately target master regulator proteins in about a third of the patients. They'll also seek to identify additional drugs that might help patients lacking those specific master regulators. And in another clinical trial for pancreatic cancer patients, sponsored by the Lustgarten Foundation, his lab will work with CUIMC pancreatic-cancer specialist Kenneth Olive and oncologists in the center to test drugs predicted to be effective by OncoTreat, the only test designed to predict drugs matched to 10 aggressive tumors in patients using the tumor RNA profile. Those tumors include ovarian, breast, pancreas, prostate, bladder and lung cancer, as well as meningioma, sarcoma, glioblastoma and GEP-NETs. The test was co-developed by DarwinHealth, a precision-oncology company that grew out of the Califano Lab and Columbia's Department of Pathology and Cell Biology.
Onco Treat, which received Clinical Laboratory Improvement Amendments (CLIA) certification by the New York State Department of Health, is also being tested in six clinical trials, including an innovative RNA-based "N of 1" study at CUIMC. That study focuses on patients who have rapidly progressing malignancies that have failed to respond to conventional or targeted therapies. The goal of the study is to identify and provide cancer patients with more effective and personalized therapies. And on Dec. 18, 2018, the Xiamen Encheng Group Co., a pharmaceutical services and precision medicine-focused company located in Xiamen City, China, announced that it has entered into a strategic partnership with DarwinHealth. Specifically, the Encheng Group will exclusively license the DarwinOncoTarget and DarwinOncoTreat tests as part of a multi-year agreement that will grant Encheng the rights to deploy, evaluate, and market them in the Greater China region, including Hong Kong, Macao, and Taiwan.
Overall, Califano is delighted by the progress his lab has made in analyzing a tumor's regulatory networks, which he says has proved to be a highly promising method for evaluating and treating cancer, "agnostic to its mutational state and even to its typical organ by organ classification."
"Such an approach is especially promising for patients with aggressive tumors that lack actionable DNA mutations," adds Califano. "It will also help patients who fail to respond to targeted inhibitors or immune-checkpoint inhibitors or who relapse following initial response to standard drug treatment. These patients, who unfortunately represent the majority of the aggressive tumor cases, have few if any effective therapeutic options right now. We think OncoTreat will offer oncologists alternatives when they run out of approved therapies—alternatives that are not predicated on educated guesswork but on an increasingly mechanistic understanding of cancer-cell regulation and response to drugs, Ultimately, we hope to make biology a much more precise and truly predictive science for treating cancer."