Mad cow disease: A computational model reveals the mechanism of replication of prions
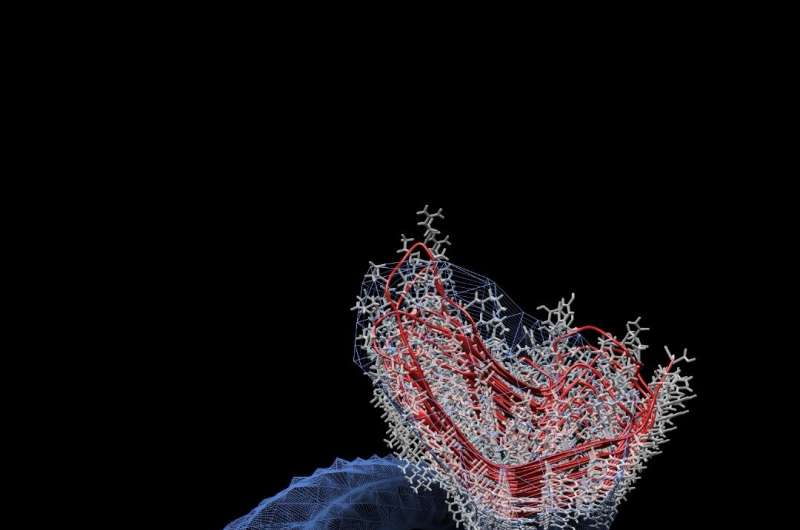
An article published in the journal PLOS Pathogens reports a realistic computational model for the structure and mechanism of replication of prions, infectious agents responsible for mad cow disease and other neurodegenerative disorders of human and animals.
The study was carried out in the Dulbecco Telethon Laboratory of Prions & Amyloids at CIBIO, led by Emiliano Biasini, University of Trento, and involved the team led by Prof. Pietro Faccioli, a physicist from the same university and affiliated with the Italian National Institute of Nuclear Physics.
Prions are unusual infectious agents made by aberrantly folded forms of a physiological protein called the cellular prion protein, or PrPC. These pathogens are known to replicate in absence of genetic material by recruiting normal PrPC molecules at the surface of cells and forcing them to change conformation and become infectious themselves. The resulting accumulation of prion particles in the nervous system lies at the root of neurodegenerative conditions known as transmissible spongiform encephalopathies, including Creutzfeldt-Jakob disease, fatal familial insomnia and Gerstmann-Sträussler-Scheinker in humans; a variety of other pathologies occur in mammals, including mad cow disease, which, in the 1990s, caused widespread epidemics in the U.K. and Europe, and several cases of cross-species transmission to humans caused by the ingestion of infected meat.
"Even though scientists have known the existence of prions since 1982, thanks to the work of Nobel laureate Stanley Prusiner, direct information regarding the structure of these non-canonical infectious agents is still lacking," says Emiliano Biasini, associate professor at the Department CIBIO, University of Trento. "In fact, their insoluble and aggregated nature hampers the use of classical high-resolution techniques for studying protein structures such as X-ray crystallography or nuclear magnetic resonance. However, such information is instrumental to rationally design drugs against these agents. In an attempt to fill this gap, we found unexpected help from a discipline usually considered far away from biology or chemistry, that is particles physics."
Telethon researchers revised previous models of prion structure and proposed a novel architecture consistent with recent experimental data. This new model allowed Pietro Faccioli's group to apply their innovative algorithms for the reliable prediction of protein conformational transitions to the prion replication mechanism "Cross-disciplinarity has been the key," explains Giovanni Spagnolli, Ph.D. student at the Department CIBIO, University of Trento, and first author of the paper. "Without the contribution of colleagues from physics, we would have never been able to afford the kind of calculation required to simulate such complex systems. For the first time, we have reconstructed a physically plausible mechanism of prion replication, which now allow us to formulate new hypotheses and design new drug discovery schemes to tackle the neurodegenerative processes unleashed by these infectious agents."
"The calculation algorithms that allowed the reconstruction of prion replication are derived from mathematical methods of theoretical physics, originally formulated to study phenomena of the subatomic world, such as the quantum tunneling effect. These mathematical methods have been adapted here to allow the simulation of complex biomolecular processes such as the folding and aggregation of proteins," says Pietro Faccioli, associate professor at the Department of Physics, University of Trento, and affiliated to the Italian National Institute for Nuclear Physics.
More information: Giovanni Spagnolli et al. Full Atomistic Model of Prion Structure and Conversion, PLOS PATHOGENS (2019). DOI: 10.1101/505271. Preprint archive: www.biorxiv.org/content/biorxi … 2/22/505271.full.pdf