Standard pathology tests outperform molecular subtyping in bladder cancer

While trying to develop a comparatively easy, inexpensive way to give physicians and their patients with bladder cancer a better idea of likely outcome and best treatment options, scientists found that sophisticated new subtyping techniques designed to do this provide no better information than long-standing pathology tests.
They looked at several sets of data on cancer specimens from patients with muscle invasive bladder cancer, a high-grade cancer associated with high mortality rates. The datasets included the one used to determine emerging molecular subtypes, and had outcome information on patients.
They consistently found that molecular subtyping of bladder tumors, which is currently being offered to patients, was outperformed by standard tests long-used by pathologists to characterize cancer as low- or high-grade and to determine the extent of its invasion into the bladder wall, surrounding fat, lymph nodes, blood vessels and beyond, they report in a study featured on the cover of the Journal of Urology.
"Muscle invasive bladder cancer is aggressive, it often has a very bad prognosis," says Dr. Vinata B. Lokeshwar, chair of the Department of Biochemistry and Molecular Biology at the Medical College of Georgia at Augusta University. "Everyone is trying to find out how to improve diagnosis, treatment and survival."
"Genetic profiling of a patient's tumor definitely has value in enabling you to discover the drivers of growth and metastasis that help direct that individual's treatment, even as it helps to identify new treatment targets," says Lokeshwar, the study's corresponding author and a member of the Georgia Cancer Center. "But using this information to subtype tumors does not appear to add diagnostic or prognostic value for patients."
Rather the investigators suggest that more study is needed before molecular subtypes are used to help guide patient care.
Evolving diagnostic approaches include compiling databanks on gene expression and mutations present in a cancer type to find patterns of gene expression that are then used to subtype tumors that "pathologically look similar" but are molecularly different. The idea is that molecular subtypes are better equipped to indicate which cancer is more or less aggressive and to help steer treatment options like whether chemotherapy before surgery to remove a diseased bladder is better.
It was RNA sequencing, or RNA-Seq, and a federal databank of genetic material from a wide range of cancers that enabled investigators from around the globe to examine gene expression in a particular tumor type, looking for common expression of some genes that correlate with a particular clinical outcome. Two subtypes, luminal, which predicts better survival, and basal, which predicts poor prognosis, were first identified for muscle invasive bladder cancer, and a total of six subtypes have now emerged. The first paper on subtypes in muscle invasive bladder cancer was published in the journal Nature in 2014.
But in their search to find a simpler, cheaper, widely available test to provide similar insight, investigators found that these emerging subtypes were outperformed by the usual clinical parameters like the tumor's grade and its spread to lymph nodes or blood vessels, Lokeshwar says.
Their work began in earnest with an exhaustive review by graduate students Daley S. Morera and Sarrah S. Lahorewala of the datasets on patients and differing classification methods used to identify the molecular subtypes.
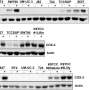
They found 11 genes that were common in all subtype classification methods. They thought, if they were going to develop a widely available test, subtyping based on these common genes might suffice. They decided to call their new subtyping panel, MCG-1.
Instead of doing RNA-Seq, which costs several thousand dollars, they used the readily available reverse transcription quantitative PCR method costing less than $10, which also looks at gene expression and is actually used to verify RNA-Seq data, Lokeshwar says.
They first looked at their own cohort of 52 patients with bladder cancer, 39 of whom had muscle invasive disease. They found MCG-1 was only 31-36% accurate at predicting important indicators like likelihood of metastasis; disease specific survival, meaning surviving bladder cancer; or overall survival, meaning survival from all causes of death from the time of cancer diagnosis or beginning of treatment until the study's end.
Recognizing that the dataset they used was comparatively small and that they did not use RNA-Seq for analysis, they then used three patient datasets from the cancer database ONCOMINE which had more patients—151 with muscle invasive bladder cancer—and also used RNA-Seq to look at gene expression.
"We found the same thing: MCG-1 could not predict disease-specific mortality," Lokeshwar says. On some patients in this dataset, information on response to chemotherapy, like commonly used cisplatin-based chemotherapy following surgical removal of the bladder, was available but subtypes could not predict chemotherapy response either, she says.
Next they looked at the dataset that has been used by a large network of investigators to identify the subtypes, The Cancer Genome Atlas, or TCGA. TCGA is a project of the National Cancer Institute and National Human Genome Research Institute that started in 2006, and has collected genetic material for 33 different cancers. The dataset includes routine pathology information on 402 specimens from patients with muscle invasive bladder cancer. It also includes these patients' overall survival and recurrence-free survival—that is when or if their cancer returned or progressed.
"Up until this point, we had been looking at patients that other groups had not looked at," Lahorewala says.
In this dataset MCG-1 predicted overall survival similar to findings reported from subtypes in several high profile publications.
"We were intrigued why MCG-1 could not predict anything in our cohort or the ONCOMINE dataset but predicted overall survival in the TCGA dataset," says Morera.
So they looked again at the 402 patients whose specimens were in the dataset and found that 21 patients' tumors were actually low-grade. Patients with low-grade tumors have higher survivability and a better prognosis than patients with high-grade muscle invasive disease.
When they removed the low-grade cases from the TCGA dataset, MCG-1 accurately predicted essentially nothing, not even overall survival. Then they included some patients with low-grade tumors into their own dataset, which they had looked at originally, and MCG-1 was now able to predict metastasis and disease specific survival, the investigators say.
All the existing subtypes are categorized as bad or better based on the cancer prognosis, the investigators say. The presence of the low-grade tumors in the classification of subtypes skewed the data to make it look like subtypes were predicting overall survival when really it was the grade of the cancer itself that was predictive.
"As investigators the first thing we did was to question our findings, since the results were so different than those reported by others," says Lokeshwar.
With the help of MCG biostatistician and coauthor Dr. Santu Ghosh, they also went back and looked at the same patients in the TCGA datasets and the subtypes they had been assigned by three different classification methods established by a network of bladder cancer researchers.
"Even with these established classification methods, the subtypes were accurate only about 50% of the time in predicting patients' overall survival. And once again, routine pathology parameters like invasion into lymph nodes or blood vessels were more accurate than the established subtypes in predicting patients' prognosis," says Lahorewala.
A recent study by investigators at Sweden's Lund University published in the journal Urologic Oncology supports the MCG investigators' findings. Their study of 519 patients who had their bladders removed because of bladder cancer found subtypes were not associated with cancer-specific survival.
Part of the problem with subtyping may be the inherent heterogeneity of tumors, says Morera. There is tremendous heterogeneity in the gene expression of tumors, even among the same tumor type, like bladder cancer, and within different parts of the same tumor as well. Furthermore, this pattern of heterogeneity can change both during tumor growth and treatment.
"Just because it's bladder cancer does not mean it's the same in all patients. We know that tumors are very dynamic and so there is heterogeneity," Lokeshwar says.
"Because there is heterogeneity, there could be problems when you want to categorize a tumor into a single subtype," says Morera.
As the name indicates, muscle invasive bladder cancer has already spread from the lining of the sac-like organ to its muscular wall. High-grade tumors, if not detected early, will spread into bladder muscle, whereas, low-grade tumors are rarely invasive. Painless blood in the urine is the most common sign of bladder cancer although only a small percentage of the individuals with it have cancer. Smoking is the major risk factor for bladder cancer.
More information: Daley S. Morera et al, Clinical Parameters Outperform Molecular Subtypes for Predicting Outcome in Bladder Cancer: Results from Multiple Cohorts, Including TCGA, Journal of Urology (2019). DOI: 10.1097/JU.0000000000000351