CRISPR screening tool identifies new drug target for leukemia
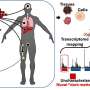
A CRISPR screening tool identified a new therapeutic target to treat acute myeloid leukemia (AML) that has the potential to leave patients with fewer side effects than current approaches, according to a new study from Penn Medicine published online in Molecular Cell. The target, known as ZMYND8, isn't a mutated gene, rather an epigenetic regulatory protein that cancer cells need to control gene expression crucial for them to stay alive and grow.
"We've discovered that cancer cells in patients with AML rely heavily on ZMYND8, and thanks to a sophisticated CRISPR-based screening approach, we pinpointed the exact 'druggable pocket' to target," said senior author Junwei Shi, Ph.D., an assistant professor of Cancer Biology in the Perelman School of Medicine at the University of Pennsylvania, and member of the Penn Epigenetics Institute and Abramson Family Cancer Research Institute.
"The findings suggest that delivering drug inhibitors against ZMYND8 could disrupt the AML vulnerable gene regulation circuits," added Zhendong Cao, a Ph.D. student investigator in Shi's lab. "It's an opportunity to develop better precision medicine compounds than current treatments to treat this blood cancer—which we are currently working on right now."
AML affects more than 20,000 patients a year, including both children and adults, and has a five-year survival rate of just 27 percent for people over 20. The standard of care includes chemotherapy; however, not all patients respond, so newer approaches are needed to expand options and improve survival.
CRISPR has allowed scientists to not only modify genes with more ease and less cost than previous approaches, but also enabled them to simultaneously screen for thousands of specific functional protein domains with high potential for therapeutic targeting.
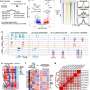
The researchers used CRISPR to precisely disrupt the domain function of proteins in cancer cells, map their molecular functions, and modify them to use in mouse models. They found that inhibiting the epigenetic reader function of ZMYND8 in mice left them with smaller tumors and better survival.
The researchers also found a biomarker—the expression level or the epigenetic status of the gene IRF8 from AML cells—to predict the sensitivity of cancer cells to a ZMYND8 inhibitor. Furthermore, the researchers validated the high expression of IRF8 and presence of IRF8 enhancer DNA element using blood samples from patients treated at Penn Medicine to support their finding.
"Many genetic and epigenetic alterations have been identified in cancer but few are actionable targets," said co-author Shelley L. Berger, Ph.D., the Daniel S. Och University Professor in the departments of Cell and Developmental Biology and Genetics in the Perelman School of Medicine and director of the Penn Epigenetics Institute. "CRISPR revealed here, for the time, an unexpected epigenetic-linked molecular circuity that AML is dependent on, and one that we can potentially manipulate. It opens a new door toward better treatments for these patients using next-generation epigenetic inhibitors."
More information: Zhendong Cao et al, ZMYND8-regulated IRF8 transcription axis is an acute myeloid leukemia dependency, Molecular Cell (2021). DOI: 10.1016/j.molcel.2021.07.018