Biomarker discovery makes early detection of high-risk COVID-19 patients possible
Researchers have discovered a biomarker that could assist in the early identification of people at high risk of developing severe COVID-19.
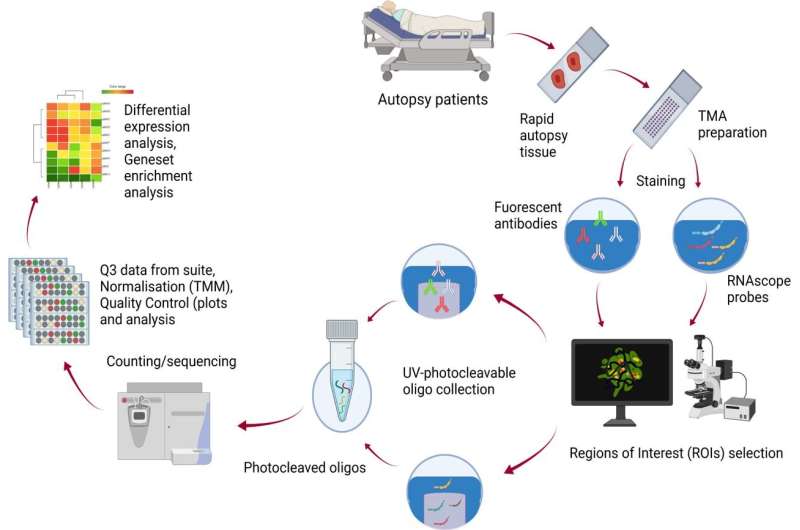
Led by computational researchers from WEHI, in collaboration with The University of Queensland, Queensland University of Technology and Hospital Marcelino Champagnat in Brazil, the study used advanced spatial transcriptomic techniques to screen for genes associated with excessive inflammation in the lungs, a key indicator of severe COVID-19.
Recently published in the European Respiratory Journal the findings have the potential to revolutionize the way patients are treated and alleviate pressure on the nation's healthcare system.
The researchers are now participating in an international effort to translate this research into a diagnostic test to identify patients at high-risk of severe COVID-19 during the early stages of their infection, to better target health-care intervention and prevent ICU admissions associated with severe disease.
Early marker of severe disease
The research team collected samples from 30 patients across three groups: 10 patients with COVID-19, 10 with H1N1 influenza and 10 uninfected.
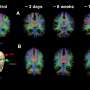
Dr. Chin Wee Tan said the team was able to generate a gene transcriptional landscape showing how different parts of the lung are impacted in each scenario.
"Only a limited number of biomarkers were found to be significantly upregulated in the lungs of COVID-19 patients, compared to patients with influenza. The presence of the IFI27 gene was a reliable prediction of severe lung inflammation," Dr. Tan said.
"Our multi-cohort follow up study, has shown that expression of the IFI27 biomarker in COVID-19 patients can predict disease progression and is strongly associated with disease severity."
Associate Professor Melissa Davis said the discovery would pave the way for a diagnostic test to be developed, so patients who were at high-risk of severe COVID-19 could be triaged and treated early.
"When a patient presents to a clinic, we could assess how severe their symptoms will become by measuring the IF127 levels in the blood—regardless of the symptoms they're presenting," Associate Professor Davis said. "Such early detection of high-risk patients will help improve patient triage; not only during the current pandemic, but potentially future viral pandemics as well."
New tools for future pandemics
Associate Professor Davis said the new Nanostring GeoMX technology and the analysis approaches developed by her team, had been crucial in better understanding how COVID-19 affects lung tissue.
"We used a technique called spatial transcriptomics, which enables us to add extra dimensions to the information we can gather from samples," she said.
"It can account for location and tissue diversity in a way that is not possible with other technologies. In this case, it provides a deeper picture of cellular changes driven by viral infection. It allowed us to define the host response to the virus and the spatial relationships between the lung and virally infected cells."
More information: Arutha Kulasinghe et al, Profiling of lung SARS-CoV-2 and influenza virus infection dissects virus-specific host responses and gene signatures, European Respiratory Journal (2021). DOI: 10.1183/13993003.01881-2021