Potential identification of SARS-CoV-2 human emergence and new COVID-19 therapeutics

Through collaboration that incorporates the use of computational modeling, data, and virology, a group of Virginia Tech researchers tackles the latest questions surrounding COVID-19. The group's project in COVID-19 human adaptation and transmission, "A selective sweep in the Spike gene has driven SARS-CoV-2 human adaptation," was published in Cell. Now, the team is building on that research for therapeutic discovery.
In 2020, prior to the beginning of the pandemic, Anne M. Brown, assistant professor and University Libraries' science informatics consultant and health analytics coordinator, and James Weger-Lucarelli, assistant professor in the Virginia-Maryland College of Veterinary Medicine's Department of Biomedical Sciences and Pathobiology, were analyzing mutations in the Mayaro virus, which causes severe arthritis in humans.
"We were interested in understanding how Mayaro virus might adapt to mosquitoes that commonly bite humans and are abundant in tropical areas," said Weger-Lucarelli. "Dr. Brown is capable of analyzing how viral mutations impact the structure of proteins, which might impact their ability to infect their target cells. When the COVID-19 pandemic hit the U.S., we saw a natural extension of our previous work with SARS-CoV-2."
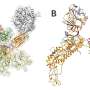
In the meantime, Weger-Lucarelli and his colleague Pawel Michalak, from the Edward Via College of Osteopathic Medicine in Monroe, Louisiana, were working on advanced sequencing methods that identified selective sweep mutations in SARS-CoV-2 that could have led to human adaptation and enabled sustained human-to-human transmission. Upon the discovery of several mutations of interest, they reached out to Brown to tap her expertise as a computational biochemist and rationalize the impact of mutations on the spike (S) protein, which mediates receptor binding and fusion to ACE2 and causes human infection. Both Weger-Lucarelli and Michalak served as corresponding authors on the work.
"Dr. Weger-Lucarelli led the project to identify potential mutations using cutting-edge techniques in the lab but needed to understand the biochemical and structural impact of such a change—to rationalize it at the atomistic level and incorporate the role of glycans in this discovery," said Brown. "We analyzed the structural data provided, connected the emerging influence of glycans on spike dynamics, and helped connect the dots."
They worked through possible structural mutations through data modeling and highlighted a small change that could have driven human adaptation, essentially what changed to make it harmful and transmissible to humans.
"This collaboration is unique because of how different our expertise is," said Weger-Lucarelli. "Dr. Brown is a molecular modeler and bioinformatician, and I am a virologist. We have been able to combine our different skill sets productively."
Amanda Sharp, a graduate student in genetics, bioinformatics, and computational biology, worked with Brown in mapping the mutation and finding ways to clearly and accurately display the complex data visually. Additionally, Xiaofeng Wang, associate professor in the School of Plant and Environmental Sciences in the College of Agriculture and Life Sciences, optimized methods for manipulating the SARS-CoV-2 genome and is an author on this work.
"This project really highlighted how we all bring talents to the table, work together, and can do some very impactful science together," said Brown.
This research gives a better understanding of the genetic changes that were necessary for SARS-CoV-2 to sustain transmission in humans following its emergence from animal reservoirs. According to Weger-Lucarelli, given the importance of this mutation, this part of the virus's protein can be targeted by antivirals or vaccines to treat the disease.
Brown and Weger-Lucarelli are continuing the collaboration and pursuing funding to study recent mutations in SARS-CoV-2 in humans that are helping it to further adapt to human transmission. They were recently awarded a Virginia Tech CeZAP interdisciplinary team-building pilot grant to molecularly barcode SARS-CoV-2 to probe in vivo evolutionary dynamics. In addition, they received a Virginia Tech Data and Decisions grant to build on their previous research, expand their research team collaborators, and develop antivirals to fight COVID-19.
"We have assembled a collaborative group that combines Dr. Brown's and my expertise, along with excellent computational and medicinal chemistry collaborators," added Weger-Lucarelli.
While working on the evolutionary emergence of SARS-CoV-2, Brown and Weger-Lucarelli were also beginning a collaboration with Sanket Deshmukh, assistant professor in chemical engineering in the College of Engineering, and Andrew Lowell, assistant professor of chemistry in the College of Science, to investigate new routes to repurpose drugs that could target the SARS-CoV-2 Mpro protein, a major target for antiviral therapeutics for COVD-19.
According to the group, remdesivir is the only antiviral currently approved for use to treat COVID-19. With the large amount of data available about existing drugs and natural products, it's advantageous to begin drug discovery by using existing compounds as starting molecules. Virginia Tech awarded the group a Data and Decisions seed grant to expand the collaboration to computationally predict compounds, validate their mechanism of action, and use data-driven approaches to functionalize new CoV-2 specific compounds.
The research group's vision is to use recent advancements in relative free energy calculations and computing resources to rationally design and quantitatively predict drugs that could make a difference in treatment of the virus. They will use advanced algorithms and data modeling to predict which compounds best target the aspect that makes the virus replicate. Then, they will test those first in the lab. This will make the drug discovery process more efficient and effective.
"To me," said Brown, "this is how science should work, truly working together to bring more of these pieces of the puzzle together more rapidly in order to have the greatest impact, and move the needle forward on the knowledge we have on these biological questions."
More information: Lin Kang et al, A selective sweep in the Spike gene has driven SARS-CoV-2 human adaptation, Cell (2021). DOI: 10.1016/j.cell.2021.07.007