New biopsy technology for analyzing multiple tumor tissue biomarkers
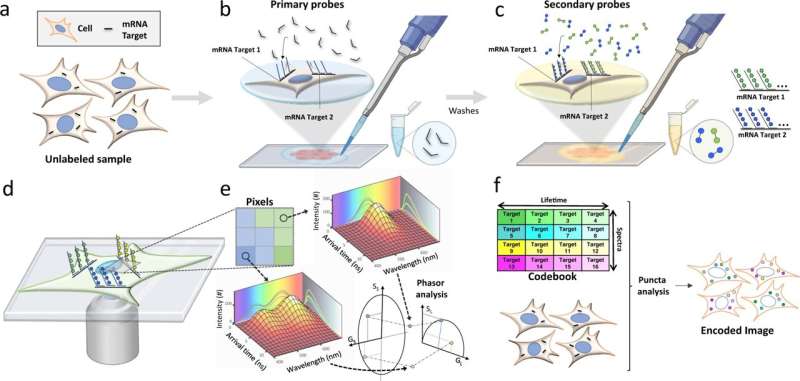
A team led by University of California, Irvine researchers has developed a new biopsy technology that can profile multiple tumor microenvironment biomarkers simultaneously, revealing cellular spatial organization and interactions that will help advance personalized disease diagnosis and treatment. Current single-biomarker biopsies lack the ability to analyze many different markers and often fail to predict patient outcomes.
Called the multi-omic, single-scan assay with integrated combinatorial analysis, the fluorescence imaging-based technology can spatially profile a large number of mRNA and protein markers in cells and tissues, including clinical tumor tissues. A study published today in Nature Communications shows that MOSAICA enables direct, highly multiplexed biomarker profiling in a 3D spatial context using a single round of staining and imaging instead of the repeated processing steps typically needed in conventional methods.
Clinicians and scientists will now have a holistic view of the different immune and cancer cell types in tumor tissues, providing greater insight for determining patient prognosis and treatment.
"Spatial biology is a new science frontier and mapping out each cell and its function in the body at both the molecular and tissue level is fundamental to understanding disease and developing precision diagnostics and therapeutics," said Weian Zhao, Ph.D., UCI professor of pharmaceutical sciences and study co-corresponding author. "Many cancer immunotherapeutics, including immune checkpoint inhibitors, don't work and scientists realized that was because of the spatial organization of all the tumor tissue cell types, which dictates drug efficacy. The MOSAICA can characterize the spatial cellular compositions and interactions in the tumor immune microenvironment in biopsies to inform personalized diagnosis and treatment."
More information: Tam Vu et al, Spatial transcriptomics using combinatorial fluorescence spectral and lifetime encoding, imaging and analysis, Nature Communications (2022). DOI: 10.1038/s41467-021-27798-0