New insights into transmissibility of COVID-19 variants and susceptibility to infection
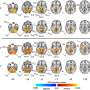
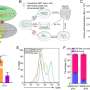
![Sensitive single-molecule detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomic RNA in infected cells. (A) Schematic illustration of single-molecule fluorescence in situ hybridisation (smFISH) for detecting SARS-CoV-2 positive strand genomic RNA (+gRNA) within infected cells. (B) Reference spatial profile of a diffraction-limited +ORF1a smFISH spot. The calibration bar represents relative fluorescence intensity (top). Frequency distribution of smFISH spot intensities, exhibiting a unimodal distribution (bottom). (C) Assessment of smFISH detection sensitivity by a dual-color co-detection method. Maximum intensity projected images and corresponding FISH-quant spot detection views of ODD and EVEN probe sets are shown. Scale bar = 5 µm. Density histogram of nearest-neighbor distance from one spectral channel to another (top). Vertical line indicates 300 nm distance. Percentage overlap between spots detected by ODD and EVEN split probes, calculated bidirectionally (bottom). (D) Heatmap of probe sequence alignment against various Coronaviridae and host transcriptomes. Each column represents individual 20 nt + ORF1a probe sequences. The minimum edit distance represents mismatch scores, where ‘0’ indicates a perfect match. Melting temperatures of each probe at the smFISH hybridisation condition are shown. (E) smFISH against +ORF1a in SARS-CoV-2-infected formalin-fixed paraffin-embedded (FFPE) lung tissue from Golden Syrian hamster at 4 days post infection. Hamsters were infected intranasally with 5 × 104 plaque-forming unit (PFU) of SARS-CoV-2 BVICO1. At necropsy, lung samples were fixed in 10% buffered formalin and embedded in paraffin wax. Red arrows in magnified panels indicate single-molecule RNA spots. Scale bars = 1000, 10, or 2 µm. (F) Experimental design for visualizing SARS-CoV-2 gRNA with smFISH at different timepoints after infection of Vero E6 cells. Cells were seeded on cover-glass and 24 hr later inoculated with SARS-CoV-2 (Victoria [VIC] strain at multiplicity of infection [MOI] 1) for 2 hr. Non-internalized viruses were removed by trypsin digestion and cells fixed at the timepoints shown. Representative 4 µm maximum intensity projection confocal images are shown. The calibration bar labeled with the symbol ‘#’ is used to display wider dynamic contrast range. Magnified view of insets in the upper panels is shown in lower panels. Scale bars = 10 µm or 2 µm. Credit: DOI: 10.7554/eLife.74153 New insights into transmissibility of COVID-19 variants and susceptibility to infection](https://scx1.b-cdn.net/csz/news/800a/2022/new-insights-into-tran.jpg)
Research led by scientists at the MRC-University of Glasgow Centre for Virus Research and the University of Oxford, in collaboration with UKHSA and Brighton NHS have revealed new insights into transmissibility of variants and susceptibility to infection.
RNA, a central molecule to the SARS-CoV-2 life cycle, encodes the genetic information required to generate new viral proteins and infectious particles. Therefore, it is vital to understand how SARS-CoV-2 RNA multiplies in infected cells. This new study, published in eLife, has developed a new approach to quantify each individual molecule of SARS-CoV-2 RNA within infected cells with greater precision and sensitivity than previously possible. This approach allowed the researchers to study the very first stages of infection and to estimate the speed in which different variants of concern of SARS-CoV-2 replicates in cells in culture.
SARS-CoV-2, the virus that causes COVID-19, was identified at the end of 2019 in the city of Wuhan. Since then, the virus has spread across all continents causing a pandemic with millions of deaths and leading to a global health and socioeconomic crisis. Thanks to the extensive effort of researchers and health workers, our knowledge on this pathogen accumulated at unprecedented rate, leading to successful vaccine programs that have reduced the burden of COVID-19. However, the emergence of variants with increased infectivity and potential to evade protective antibodies produced by vaccination, is a major current concern.
The study showed that the alpha variant, detected months ago in the UK, has a slower replication kinetics than the original virus, suggesting that SARS-CoV-2 variants of concern might not only differ in their ability to enter the cells, but also in the speed in which they multiply their RNA.
Dr. Alfredo Castello, said: "Most of the attention has been centered on the mutations detected in the virus spike protein, as they can influence the ability of the virus to enter the cell or to escape vaccine induced or natural antibody responses. However, we should not ignore that there are many other mutations occurring in other viral proteins that can influence virus growth and transmission."
This study pioneered the use of single molecule analysis to study SARS-CoV-2 replication in cells. Applying this approach to other variants of concern such as OMICRON will shed light into their replication dynamics and how these may affect clinical outcomes.
Professor Ilan Davis said: "Most of the available techniques to study viral replication rely on studies of cell populations, providing data that is an average between all cells within the culture or tissue. While useful, it does not allow us to see differences between cells or to study the SARS-CoV-2 life cycle with enough detail.
"Our approach surpasses these 'in bulk' techniques because it provides accurate information for each individual cell from the very first time point of infection to the point at which the cell is completely overwhelmed with viral RNA. We were also able to show that the technique works in sections from infected lungs of animals, which opens up possibilities in the future to study samples from infected patients."
Thanks to this technical tour-the-force and close cooperation between a diverse team of researchers, this study revealed that SARS-CoV-2 infection varies across cells, with the majority of cells showing low levels of viral RNAs. However, in a minority (10%) of cells contained extremely high levels of virus RNA.
Jeff Lee, co-author of the study, said: "This level of cell heterogeneity is surprising because these cells were clones. In other words, they are in principle near-identical. "I would expect similar or greater variation between cells in complex tissues, which are heterogenous and contain many different cell types."
Peter Wing, co-author, said: "The high variation in SARS-CoV-2 replication in clonal cell lines implies that the cellular state, such as the cell cycle or the response to stress, may define the speed in which the virus replicates. It would be of great therapeutic interest to discover the determinants of cell susceptibility to infection."
The study also revealed that SARS-CoV-2 RNAs are long-lived within infected cells.
Professor Jane McKeating, said: "When we treated the cells with remdesivir to inhibit viral replication, we observed that the viral RNA persisted for a long time, being detectable for many hours after the treatment. "This could explain why SARS-CoV-2 RNA can be detected in some patients long after the clinical symptoms have resolved."
The paper, "Absolute quantitation of individual SARS-CoV-2 RNA molecules provides a new paradigm for infection dynamics and variant differences," is published in eLife.
More information: Jeffrey Y Lee et al, Absolute quantitation of individual SARS-CoV-2 RNA molecules provides a new paradigm for infection dynamics and variant differences, eLife (2022). DOI: 10.7554/eLife.74153