CryoEM unlocks understanding of COVID-19 evolution
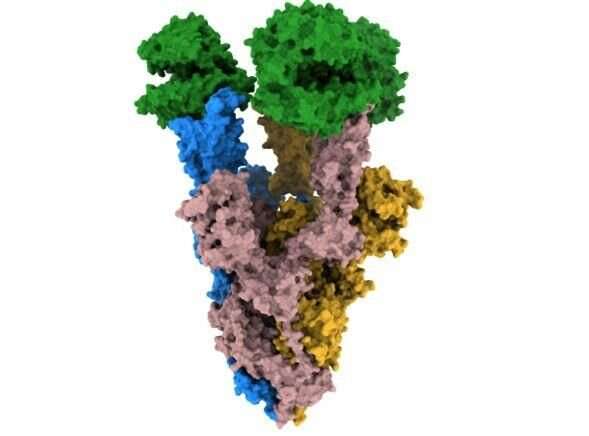
Researchers at the Francis Crick Institute have identified structural changes in COVID-19 variants which provide clues to how the virus evolved to have greater levels of infectivity.
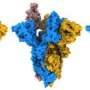
Throughout the COVID-19 pandemic, SARS-CoV-2 has evolved variants with differences in the structure of the virus spike protein. Changes to this spike affect how the virus binds to and infects cells, which is part of what controls infectivity.
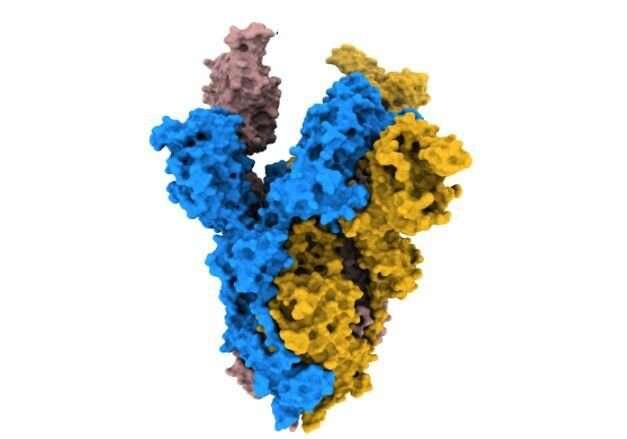
In their study, published in Nature Communications today (Friday 4 March), the scientists used high resolution cryo-electron microscopy to identify structural changes in the spike protein of the alpha and beta variants, which emerged during the pandemic in 2020.
They found a change in the alpha spike means it is more stable when it binds to the human cell receptor, ACE2, in comparison to the original strain of the virus. This increased stability likely makes the process of the virus binding to and infecting the cell more effective for the virus.
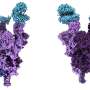
The beta spike has a different substitution, K417N, which the authors propose is crucial to arrange the spike in a more open shape, which primes the spike for binding to the cell receptor.
These two changes, which cause increased stability and a more open spike formation are also present in the omicron variant.
Antoni Wrobel, co-lead author and postdoctoral training fellow in the Structural Biology of Disease Processes Laboratory at the Crick, says: "We're aiming to understand the stages of evolution this virus has gone through, which is important as some of the changes we've seen in the alpha and beta variants have been observed in subsequent variants and might be present in future ones."
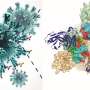
The researchers also found that both variants have the same substitution, called N501Y, in their receptor-binding domains, which is where the virus binds to human cells. When this substitution is present alongside another, called D614G, it causes the virus to bind more strongly to cells compared to the original virus. For alpha the binding was six times stronger and for beta it was two times stronger.
This observation might explain why later variants evolved from an earlier variant with the D614G substitution rather than through a different evolutionary path.
Co-lead author and postdoctoral training fellow in the Structural Biology of Disease Processes Laboratory at the Crick, Donald Benton explains: "Our work proposes three general mechanisms through which the spike has evolved in the human population. We can see how key amino acid substitutions have led to the spike protein developing increased stability, a more open structure, and the ability to form stronger bonds with human cell receptors. These changes help the virus achieve greater levels of infectivity, that is its ability to establish an infection in a host."
Steve Gamblin, author and group leader of the Structural Biology of Disease Processes Laboratory at the Crick, says: "The structure of SARS-CoV-2 has changed repeatedly as the virus has evolved, with changes increasing how effective the virus is at infecting cells. Cryo-electron microscopy provides a valuable tool to explore these changes and help us better understand how the genetic changes correspond with changes in spike structure and receptor binding."
Researchers across the Crick are continuing to study SARS-CoV-2, including to understand how it affects the immune system, and how much protection the vaccines provide.
More information: Antoni G. Wrobel et al, Evolution of the SARS-CoV-2 spike protein in the human host, Nature Communications (2022). DOI: 10.1038/s41467-022-28768-w. doi.org/10.1038/s41467-022-28768-w