Study zeroes in on genes involved in Crohn's disease

An international consortium of researchers has identified genetic variants in 10 genes that elevate a person's susceptibility to Crohn's disease, a form of inflammatory bowel disease.
Led by researchers from the Wellcome Sanger Institute and the Broad Institute of MIT and Harvard, the study is the largest to date to focus on rare variants associated with Crohn's disease and is published today in Nature Genetics. These discoveries highlight the causal role of mesenchymal cells in intestinal inflammation, helping to zero in on the genetic roots of inflammatory bowel disease and providing better data with which to develop the next generation of treatments.
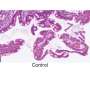
Crohn's disease (CD) is a debilitating condition characterized by chronic inflammation of the gastrointestinal tract. The causes of the disease are poorly understood, but it is believed to be triggered by a hyperactive immune response against gut bacteria in genetically susceptible individuals. Though drugs are available that improve symptoms for many patients, there is no cure and relapsing bouts of severe illness are common.
With a few rare exceptions, there is no single genetic cause of CD. Environment, diet and genetic variation collectively shape an individual's risk of disease. Previous genome-wide association studies (GWAS) have identified around 250 regions of the genome that influence an individual's susceptibility to CD. Unfortunately, GWAS studies are somewhat restricted to testing sites in the human genome that frequently vary between individuals.
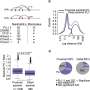
In this study, researchers at the Wellcome Sanger Institute and the Broad Institute set out to identify rare genetic variants within protein-coding genes that are associated with Crohn's disease susceptibility. They performed exome sequencing on around 30,000 patients with Crohn's and compared these to exome sequences from around 80,000 individuals without the condition.
This identified genetic variation within six genes in regions of the genome that had not been previously connected to Crohn's disease. Several of these genes are known to play important roles in a type of stem cell in the gut called mesenchymal cells, suggesting that disruption of these cells contributes to the initiation and maintenance of intestinal inflammation.
Dr. Aleksejs Sazonovs, a first author of the study from the Wellcome Sanger Institute, said, "Most humans will have some of the genetic variants that increase susceptibility to inflammatory bowel disease because they're so common. These common variants may increase a person's risk by 10%, for example, but this increased risk doesn't necessarily lead to disease. But some rare variants can make someone four or five times more likely to develop inflammatory bowel disease, so it's especially important to locate these and understand the biological processes they disrupt."
The remaining four remaining genes identified by the team reside within regions of the genome previously associated with IBD via GWAS. Unfortunately, the common genetic variants these GWAS associated with disease lie outside of protein coding genes, making it challenging to draw insights into disease biology. The identification of the particular genes in these regions that are underpinning susceptibility to Crohn's disease ends this challenge and the biological pathways in which these genes operate can now be considered for pharmaceutical interventions.
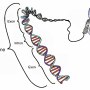
One rare variant, in the TAGAP gene, decreases a person's likelihood of developing the disease. Such variants, called "protective mutations," are appealing to researchers in part because they suggest that a certain gene may be disabled without adverse side effects in people. Drugs that mimic the mutation, such as by disabling the protein the gene encodes, could confer some of the same protection in patients.
Dr. Mark Daly, a senior author of the study from the Broad Institute of MIT and Harvard, said, "In thinking about how to develop new therapies, it's critical that we can pinpoint the specific genetic variants that increase or decrease a person's risk. When we discover a disease association to a genetic variant within a gene, we can start running experiments the next day to figure out what the variant, the gene, and the protein it encodes is doing to influence disease risk. This puts us on a dramatically faster track for converting those observations into a therapeutic hypothesis."
The next step will be to extend the approach to ulcerative colitis and increase the scale of sampling, in the hope of locating all variants and genes involved in IBD. Improved data should help to make drug discovery a more efficient and faster process, offering a glimmer of hope to those affected by the disease.
Dr. Carl Anderson, a senior author of the study from the Wellcome Sanger Institute, said, "To have the statistical power to spot the rare variants that are driving disease, these studies require tens of thousands of individuals. We need international collaborative teams, such as the International IBD Genetics Consortium, to bring together sufficient DNA samples to make this possible. We've already begun working on our next study, which will use exome sequence data from more than 650,000 individuals and give us unprecedented ability to derive insights into the aberrant biology underpinning inflammatory bowel disease."
More information: Hailiang Huang, Large-scale sequencing identifies multiple genes and rare variants associated with Crohn's disease susceptibility, Nature Genetics (2022). DOI: 10.1038/s41588-022-01156-2. www.nature.com/articles/s41588-022-01156-2