Multiomics study of nonalcoholic fatty liver disease
Scientists at deCODE genetics, a subsidiary of Amgen, published today a large genome-wide association study on nonalcoholic fatty liver disease (NAFLD) in Nature Genetics. Sequence variants that associate with NAFLD were identified, including rare, protective loss-of-function variants that point to potential drug targets. Plasma proteomic analyses provided further insight into the pathogenesis of NAFLD.
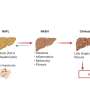
NAFLD is a growing health problem and is estimated to affect up to 25% of the world's population. Nonalcoholic fatty liver (NAFL), when over 5% of the liver is fat with no identifiable causes such as excessive alcohol consumption, is the first stage of NAFLD.
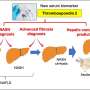
NAFL can progress to non-alcoholic steato-hepatitis (NASH) which can progress further into liver cirrhosis and hepatocellular carcinoma (HCC). NAFLD can be difficult to diagnose and monitor and there is currently no therapy available. The identification of potential drug targets and biomarkers is therefore of great importance.
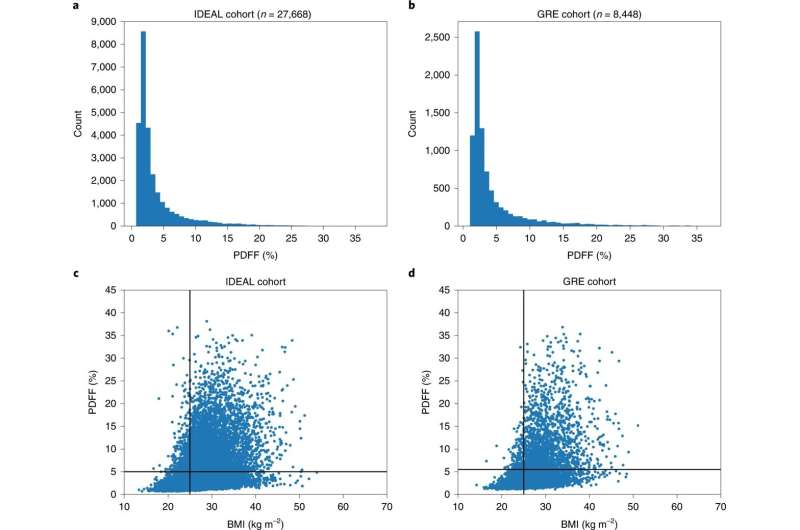
A large genome-wide association study of NAFL, liver cirrhosis and HCC was conducted and the findings integrated with expression and proteomic data. For NAFL, 9,491 clinical cases from Iceland, UK, U.S. and Finland were utilized in addition to proton density fat fraction (PDFF) extracted from 36,116 liver MRIs. Among the sequence variants discovered, in the Icelandic population, were rare, protective, predicted loss-of-function variants in MTARC1 and GPAM suggesting that inhibiting MTARC1 or GPAM could be therapeutic for NAFL or NASH.
Levels of thousands of proteins measured in plasma were analyzed, identifying potential biomarkers of disease, disease progression or target engagement and models that can discriminate between a NAFL and cirrhosis were constructed using the proteomics data. The results therefore provide a path to the development of non-invasive tools to evaluate and diagnose NAFLD.
Additionally, the pleiotropic effects of the identified variants were explored by looking at associations with 52 other phenotypes and traits. BMI is one of the most common risk factors of NAFLD and longitudinal PDFF measures suggested that carriers of p.Ile148Met, the well-know NAFLD risk variant, in PNPLA3 are more susceptible to change in BMI than non-carriers.
To date this study is one of the largest ones conducted on the genetic basis of NAFLD and the results will hopefully contribute to the development of diagnostic tools and therapies that can help patients suffering from NAFLD.
More information: Gardar Sveinbjornsson, Multiomics study of nonalcoholic fatty liver disease, Nature Genetics (2022). DOI: 10.1038/s41588-022-01199-5. www.nature.com/articles/s41588-022-01199-5