Examining resistance to immunotherapy in ovarian cancer
A team at Memorial Sloan Kettering Cancer Center (MSK) has uncovered several mechanisms of immune escape that can help explain why ovarian cancers have been resistant to immunotherapy to date.
The researchers found that ovarian cancer is even more complex than previously thought, with profound differences in immune evasion between known genomic subtypes, and even among different tumors within the same patient. They also learned that ovarian tumors develop new mutations to hide from the immune system as they spread.
The research, published December 14, 2022, in Nature, is the first study by a collaborative effort called MSK SPECTRUM, which is focused on high-grade serous ovarian cancer. This type accounts for about 80% of all ovarian cancers and is the most lethal subtype.
How cancer and immune cells interact in ovarian cancer
The immune system has a built-in braking system to keep it from overwhelming the body with a response that's too strong. This same checkpoint system can be exploited by cancer cells trying to evade immune cells. Immunotherapy drugs called checkpoint inhibitors take the brakes off the immune system and allow it to attack cancer. They've been successful in treating some advanced cancers.
"There was a lot of hope around clinical trials studying checkpoint inhibitors in ovarian cancer, but their results have been disappointing," says computational oncologist Sohrab Shah, Ph.D., one of the paper's two corresponding authors. "We set out to find out why the immune system can't be stimulated against this cancer like it can with many other diseases. What is the nature of how cancer cells and immune cells interact in ovarian cancer?"
"With this project, we studied pretreatment ovarian cancers and used the latest tools to comprehensively characterize the interaction between tumor and immune cells," adds medical oncologist and ovarian cancer expert Dmitriy Zamarin, MD, Ph.D., the other corresponding author. "We performed multiple complementary analyzes and integrated the data to better understand how ovarian cancer behaves in patients and to find answers about the lack of response to immune therapy."
People with ovarian cancer donate tumor samples
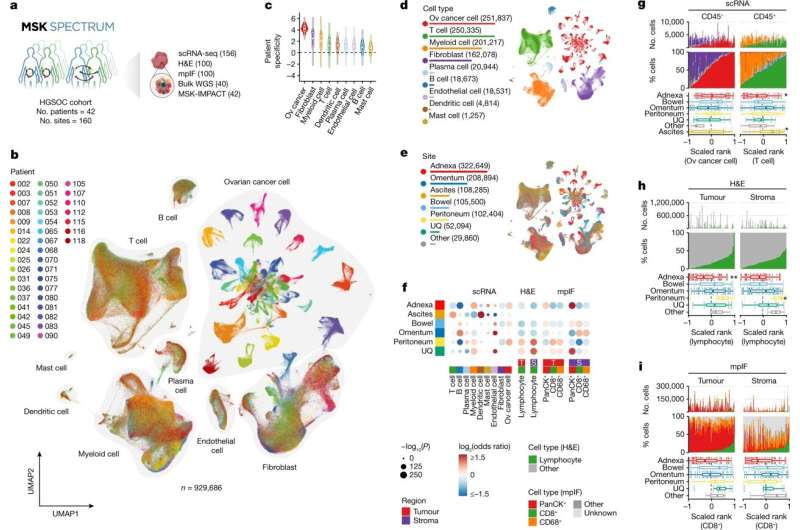
Forty-two people who had been recently diagnosed with advanced ovarian cancer and seen at MSK's gynecologic oncology surgery clinics agreed to donate tissue for this research. All the patients had high-grade serous ovarian cancer. During surgery to remove their tumors, the researchers collected samples not only from the original tumors in the ovaries and fallopian tubes but also from the areas where the cancer had metastasized (spread) in the patients' bodies. This resulted in a total of 160 unique tumors available to study.
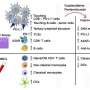
The team performed "multimodal" analyzes on the patient samples, including:
- Whole genome sequencing
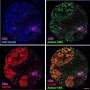
- Single-cell RNA sequencing, which enables investigators to determine the genes that are being expressed, or "turned on," in different cells, in the various contexts of where each cancer cell is in the body. Learning how cancer cells behave in their "microenvironment"—the collection of all the cell types that make up a tumor—is key to understanding why immune cells are not able to recognize cancer cells.
- Multiplexed immunofluorescence, a type of imaging that simultaneously identifies specific proteins and immune cell types in tissue samples
- Digital pathology scans
How tumors develop the ability to hide
Ovarian cancer is generally divided into subtypes based on its various gene mutations. One of the key findings was that the nature of immune recognition could be attributed to these genomic subtypes. This, in turn, could lead to improved personalized approaches to therapy for patients.
But even within the same subtype, the researchers found that there may be profound differences among tumors, suggesting they are even more complicated than previously believed. For example, some tumors within the same patient appeared to be recognized by the immune system, while others remained hidden.
"Ovarian tumors don't always respond to treatments the way we would expect them to, and such differences among the different tumors of the same patient make it challenging to identify a marker that could predict the response to therapies," says Dr. Zamarin, who has led many clinical trials for the disease.
One significant discovery reported in the Nature paper is that as ovarian cancer grows and spreads, the cancer cells develop new mutations to help them survive. Some of these mutations actually help the tumors hide from the immune system.
"There's a kind of cascading evolutionary arms race that happens as the cancer progresses," Dr. Shah says. "The original tumors appear to be recognized by the immune system, but metastatic tumors formed later, in different anatomic sites, acquire the ability to evade immune recognition."
Dr. Shah adds this discovery makes developing better early detection methods even more urgent, so that tumors can be eliminated before they find a way to evade the immune system.
Research drives the development of better ovarian cancer treatments
The research revealed a much deeper explanation for what the doctors and scientists already knew: Ovarian cancer is extremely difficult to treat. But they say their work also revealed good news that points toward possible new ways to fight the disease.
"Now that we know some of the additional mechanisms that are driving resistance to therapy, we have an opportunity to find better ways to improve treatments," Dr. Zamarin says. "The findings from this study have given us many additional avenues to pursue, not only for developing new treatments but, ultimately, also better methods for both detection and prevention."
The researchers note that the techniques developed in this study may be especially useful in studying other cancers that spread in the abdominal area, including endometrial (uterine) cancer, pancreatic cancer, and stomach cancer.
More information: Ignacio Vázquez-García et al, Ovarian cancer mutational processes drive site-specific immune evasion, Nature (2022). DOI: 10.1038/s41586-022-05496-1