This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Refined method to peer into cancer cells' inner working unlocks the interactome
For a long time, cancer genetics and epigenetics researchers have been focusing on alterations in genes and proteins to understand what makes a cell turn cancerous and become malignant. In the nucleus of the cell, there is crosstalk between highly complex gene networks, controlled by small dispersed regulatory elements through the physical contact of DNA regions.
Many cancers show alterations in this crosstalk due to, for instance, DNA gains, losses or rearrangements. While genes may be perfectly fine, being in a different position in the nucleus disrupts their ability to physically contact the regulatory elements. The study of this network of interactions, known as the interactome, is one of the most challenging and unknown fields in cancer biology.
Analyzing the interactome is not an easy task. After carefully extracting the DNA of a cancer cell, taking all possible precautions to avoid disrupting those parts of the genome that are "talking to each other," scientists stick them together using a few biochemical tricks to reveal which genes and regulatory elements are in physical contact. By comparing healthy cells to cancer cells, the researchers hope to understand the aberrant "discourse" of malignancies.
Up to now, this methodology required a fair amount of DNA to be successful and was, therefore, not useful in clinical settings due to the limited sample size obtained in a biopsy. But this may change thanks to liCHi-C—a new method developed by the 3D Chromatin Organization Lab led by Dr. Biola M. Javierre at the Josep Carreras Leukemia Research Institute,
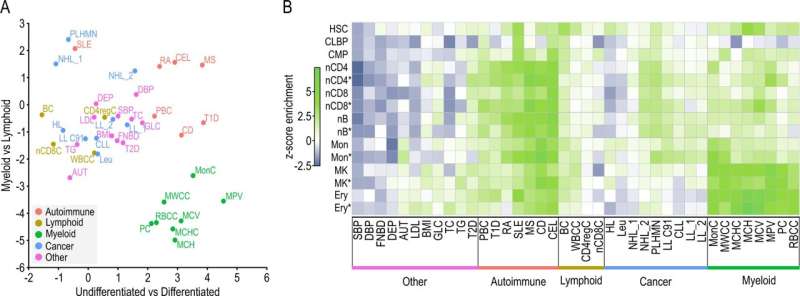
In a recent publication in Nature Communications, the researchers show that the interactome of a particular tumor can be analyzed directly from patient samples instead of from in vitro models, which is a great leap forward. This feat relies on the fact that liCHi-C, an improvement over the previous Promoter Capture Hi-C (PCHi-C) method, works with as few as 50,000 cells instead of the millions needed for other methods. This substantial reduction in sample size is possible thanks to the combination of a streamlined experimental protocol with new bioinformatic tools.
Together with colleagues from the Josep Carreras Institute, the Barcelona Supercomputing Center, Wellcome-MRC Cambridge Stem Cell Institute, Sant Joan de Déu, IDIBAPS and the Hospital Clinic among others, the researchers have been able to increase the resolution of the interactome maps in developing hematopoietic stem cells from healthy donors and cancer patients, allowing them to identify the altered networks that occur in leukemia. Also, they show how liCHi-C can be used to identify large DNA rearrangements with higher precision and to reveal the role of non-coding alteration in cancer development.
Understanding the aberrant crosstalk in cancer cells may help develop new therapies, aimed at disrupting the "toxic chatter" inside them. We are still far from the clinic, but liCHi-C is the first step toward a wider and richer description of what happens inside a cancer cell, one of the goals of biomedical research.
More information: Laureano Tomás-Daza et al, Low input capture Hi-C (liCHi-C) identifies promoter-enhancer interactions at high-resolution, Nature Communications (2023). DOI: 10.1038/s41467-023-35911-8