This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Transcriptomic analysis of human ALS skeletal muscle reveals a disease-specific pattern of dysregulated circRNAs
A new research paper was published in Aging, entitled, "Transcriptomic analysis of human ALS skeletal muscle reveals a disease-specific pattern of dysregulated circRNAs."
Circular RNAs are abundant, covalently closed transcripts that arise in cells through back-splicing and display distinct expression patterns across cells and developmental stages. While their functions are largely unknown, their intrinsic stability has made them valuable biomarkers in many diseases.
In this new study, researchers from the National Institutes of Health's National Institute on Aging, The University of Alabama at Birmingham, Birmingham Veterans Affairs Medical Center, and Cedars-Sinai Medical Center set out to examine circRNA patterns in amyotrophic lateral sclerosis (ALS). By RNA-sequencing analysis, the researchers first identified circRNAs and linear RNAs that were differentially abundant in skeletal muscle biopsies from ALS compared to normal individuals.
"By RT-qPCR analysis, we confirmed that 8 circRNAs were significantly elevated and 10 were significantly reduced in ALS, while the linear mRNA counterparts, arising from shared precursor RNAs, generally did not change," state the authors.
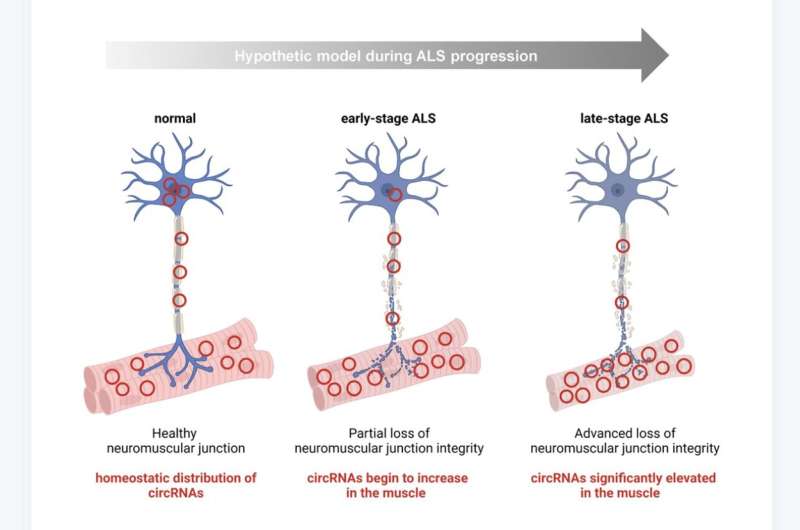
Several of these circRNAs were also differentially abundant in motor neurons derived from human induced pluripotent stem cells (iPSCs) bearing ALS mutations, and across different disease stages in skeletal muscle from a mouse model of ALS (SOD1G93A). Interestingly, a subset of the circRNAs significantly elevated in ALS muscle biopsies were significantly reduced in the spinal cord samples from ALS patients and ALS (SOD1G93A) mice. In sum, the researchers identified differentially abundant circRNAs in ALS-relevant tissues (muscle and spinal cord) that could inform about neuromuscular molecular programs in ALS and guide the development of therapies.
The researchers conclude, "As our studies advance, we will investigate the function of the most promising and abundant circRNAs, among the 18 circRNAs reported here. We are especially interested in those that appeared to be specific for ALS, as they may help to characterize disease-associated molecular pathways that could be targeted therapeutically."
More information: Dimitrios Tsitsipatis et al, Transcriptomic analysis of human ALS skeletal muscle reveals a disease-specific pattern of dysregulated circRNAs, Aging (2022). DOI: 10.18632/aging.204450