This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Automated, accurate reporting for NGS-based clonality testing
A new research paper titled "Development and implementation of an automated and highly accurate reporting process for NGS-based clonality testing" has been published in Oncotarget.
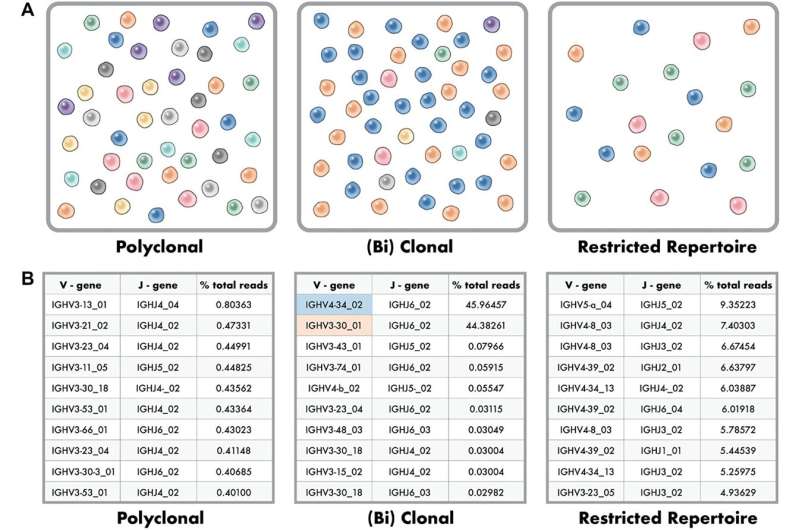
B and T cells undergo random recombination of the VH/DH/JH portions of the immunoglobulin loci (B cell) and T-cell receptors before becoming functional cells. When one V-J rearrangement is over-represented in a population of B or T cells indicating an origin from a single cell, this indicates a clonal process. Clonality aids in the diagnosis and monitoring of lymphoproliferative disorders and evaluation of disease recurrence.
In a new study, researchers Sean T. Glenn, Phillip M. Galbo Jr., Jesse D. Luce, Kiersten Marie Miles, Prashant K. Singh, Manuel J. Glynias, and Carl Morrison from Roswell Park Comprehensive Cancer Center aimed to develop objective criteria that could be automated to classify B and T cell clonality results as positive (clonal), no evidence of clonality, or invalid (failed).
"Using clinical samples with 'gold standard' clonality data obtained using PCR/CE testing, we ran NGS-based amplicon clonality assays and developed our own model for clonality reporting," the researchers explain.
To assess the performance of their model, the researchers analyzed the NGS results across other published models. Their model for clonality calling using NGS-based technology increases the assay's sensitivity, more accurately detecting clonality. In addition, they built a computational pipeline to use their model to objectively call clonality in an automated fashion.
"Collectively, the results outlined below will have a direct clinical impact by expediting the review and sign-out process for concise clonality reporting," summarize the researchers.
More information: Sean T. Glenn et al, Development and implementation of an automated and highly accurate reporting process for NGS-based clonality testing, Oncotarget (2023). DOI: 10.18632/oncotarget.28429