May 2, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Whole-genome sequencing used to track down genes behind familial glioma
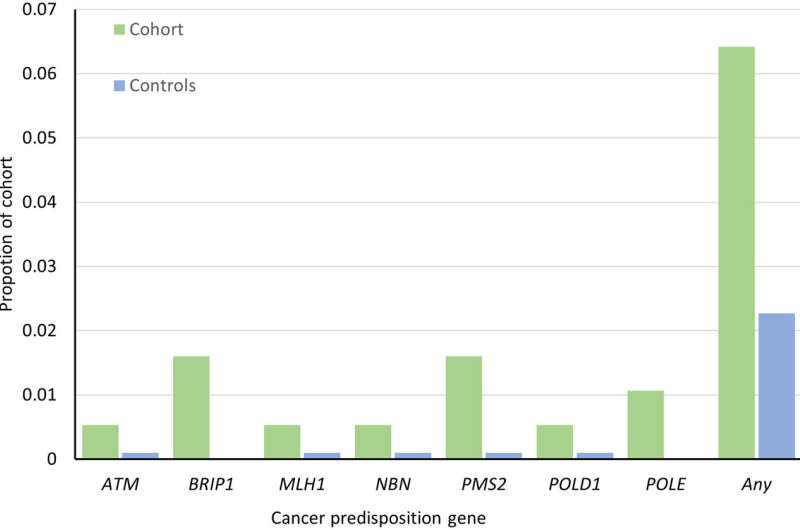
A team of gene therapists, oncologists, genetic sequencing experts and neurosurgeons affiliated with a host of institutions in the U.S. and one in Sweden has uncovered gene variants that appear to be responsible for passing on familial glioma from parent to offspring. In their study, reported in the journal Science Advances, the group sequenced the genomes of members of glioma-affected families.
Glioma is a form of cancerous brain tumor with a generally poor prognosis. Familial glioma is a subclass of glioma that persists along family lines, strongly suggesting a genetic component. In this new effort, the researchers set their sights on tracking down the specific genes responsible for passing on a propensity for developing glioma.
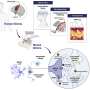
The researchers took tissue samples from 203 volunteers from 189 families with a history of glioma and performed whole-genome sequencing. They then repeated the whole exercise using tissue samples from another 122 people from 115 families. Once sequencing was completed, germline variants were compared with sequences done on more than 1,000 people who did not have a family history of glioma.
The researchers found 54 variants appearing in 28 genes that appeared to be overrepresented in families with a history of glioma. They found them in 50 of the families involved in the testing. More specifically, they found copy number changes in HERC2. They also found an overlap between genes they identified as possibly related to familial glioma and genes that have previously been associated with other kinds of cancer.
The data also showed variants they described as suspicious in some noncoding genes, which they suggest could play a role in mediating the activity of other genes. Such variants, they further note, appeared to coincide with heightened levels of transcription factor bind mutations upstream of some of the involved genes.
The researchers conclude by noting that theirs is the first study to link the HERC2 gene to predisposition to any type of cancer and the first to look at the role noncoding variants may play in glioma.
More information: Dong-Joo Choi et al, The genomic landscape of familial glioma, Science Advances (2023). DOI: 10.1126/sciadv.ade2675
© 2023 Science X Network