June 29, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Gene found that prevents most bird flu variants from jumping to humans
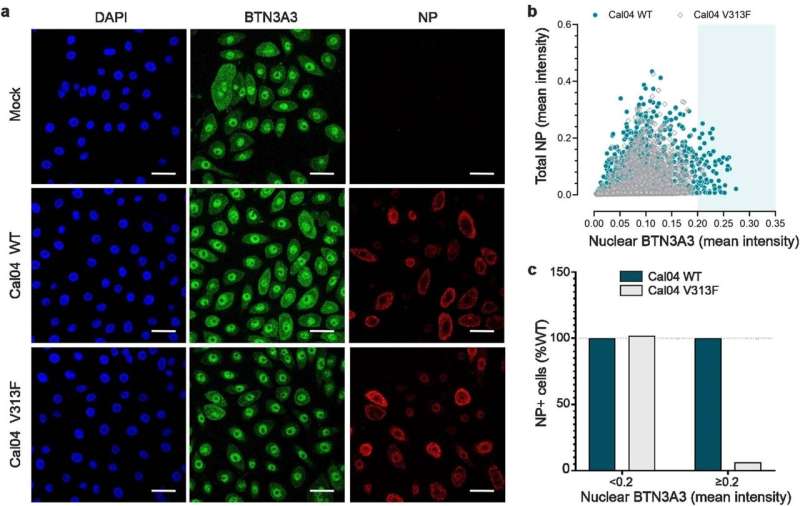
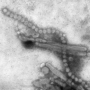
A large team of medical researchers and virus specialists at the University of Glasgow working with several colleagues from the University of Edinburgh and one from Istituto Zooprofilattico Sperimentale delle Venezie has discovered a human gene that prevents most avian flu variants from jumping to humans.
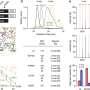
In their study, reported in the journal Nature, the group identified virus-killing properties associated with the BTN3A3 gene. Laura Graf and Peter Staeheli with the University of Freiburg, have published a News & Views piece in the same journal issue, outlining the work done by the team on this new effort.
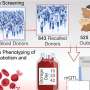
The work involved six years of screening proteins for antiviral activity in cultured human cells. The researchers discovered that most avian virus attempts to jump to human cells were blocked by the activation of the BTN3A3 gene. Once activated, an immune response ensured that the virus could not get a foothold, preventing infection. They also found that the gene is not a variant—it is present in everyone and has been in our gene pool for approximately 40 million years. A closer look showed that when viruses are identified, the gene is activated in the lungs, nose and throat, the sites most likely to be transmission areas.
The researchers also note that throughout history, viruses that have jumped from birds or swine, setting off epidemics or pandemics, have shown resistance to the activation of the BTN3A3 gene. They also note that avian flu variants are constantly evolving, which means some variants that are blocked from jumping to humans today could evolve to be resistant, allowing for such jumps in the future. The research team showed this to be the case with a virus called H7N9 that developed resistance over the years 2011 to 2013. Thus far, the strain has infected 1,570 people and killed 616.
The research team suggests their findings could simplify future work involved in assessing risk from new strains of avian flu—upon first discovery, it can be tested to see whether or not it has resistance to the protection provided by the BTN3A3 gene.
More information: Rute Maria Pinto et al, BTN3A3 evasion promotes the zoonotic potential of influenza A viruses, Nature (2023). DOI: 10.1038/s41586-023-06261-8
Laura Graf et al, A human protein that holds bird flu viruses at bay, Nature (2023). DOI: 10.1038/d41586-023-01942-w
© 2023 Science X Network