This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Researchers publish the first comprehensive, 3D analysis of the complete rhabdomyosarcoma genome
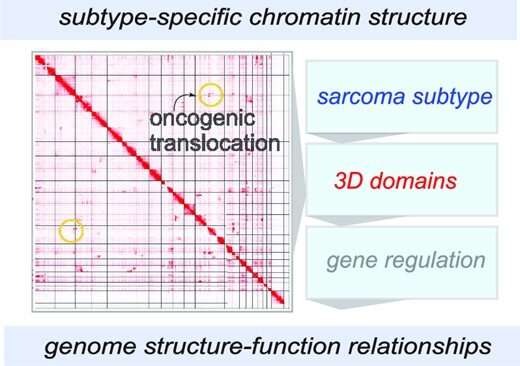
In a paper published in NAR Cancer, researchers from the Center for Childhood Cancer Research at Nationwide Children's and their collaborators report a comprehensive 3D chromatin structural analysis and characterization of rhabdomyosarcoma (RMS). RMS is a pediatric soft-tissue tumor that predominantly affects children between 0 and 4 years, but it can also affect adolescents. Survival rates vary dramatically from 10% to 90% depending on the stage and subtype.
"RMS represents an unmet medical need in pediatric cancer," says Benjamin Stanton, Ph.D., principal investigator in the Center for Childhood Cancer Research and senior author of the publication. "Few mutations have been identified in this tumor, and even then, precision medicine remains a challenge. By understanding the epigenetics—the factors affecting gene expression outside of the actual genetic code—of RMS, we can find epigenetic drivers and give the medical community new targets and vulnerabilities for precision therapy."
The Stanton Lab's latest publication describes three layers of epigenetic characterization of RMS.
"While the human genome project provided the one-dimensional sequence of base pairs, we have to realize that the human genome is three-dimensional," says Meng Wang, Ph.D., a senior bioinformatic scientist in the Center for Childhood Cancer Research and first author of the publication. "Cancers can be caused by mutations in the line of code, but they can also be caused by how different genes associate with one another in three-dimensional space."
To understand how the 3D structure of the RMS genome could contribute to tumor formation, the team began the deepest, most comprehensive analysis of the structure to date.
The team created spike-in in situ Hi-C chromatin interaction maps for the most common RMS cell lines. They uncovered common and distinct structural components in large-scale chromatin compartments, tumor-essential genes with variable topologically associating domains (TADs), and unique patterns of structural variation.
"Dr. Wang is the first to comprehensively describe the compartments and TADs of RMS," says Dr. Stanton. "This is a big step forward for the field in terms of understanding RMS and identifying possible targets for new therapies."
The team's high-depth chromatin interactivity maps and comprehensive analyses provide context for gene regulatory events and reveal functional chromatin domains in RMS.
More information: Meng Wang et al, The 3D chromatin landscape of rhabdomyosarcoma, NAR Cancer (2023). DOI: 10.1093/narcan/zcad028