August 17, 2023 feature
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Electrical detection of RNA cancer biomarkers at the single-molecule level
Early screening methods offer a promising avenue to reduce mortality rates of cancer, notably through the analysis of biomarkers in liquid biopsies. Biologists have shown how the electrical detection of nucleic acids at the single molecule level can facilitate such applications.
In a new paper published in Scientific Reports, Keshani G. Gunasinghe and a team of scientists in chemistry at the University of Massachusetts, U.S., studied the electrical detection of RNA cancer biomarkers for cancer screening applications as a proof-of-concept electrical biosensor.
Sequence-sensitive electrical conductance of the instrument allowed the discrimination of mutants from the wild-type. Their specificity showed the sensitivity of biosensors down to an individual molecule with a high signal-to-noise ratio. The outcomes pave the way to engineer miniaturized single-molecule electrical biosensors that are groundbreaking for cancer screening.
Cancer screening with scanning tunneling microscopy-assisted break junctions (STMBJ)
The World Health Organization lists cancer as a leading cause of mortality; these statistics can be evaded through the early detection of the disease via non-invasive analysis of liquid biopsies. Liquid biopsies target cancer-specific biomarkers in blood or saliva including tumor cells or circulating tumor nucleic acids such as circulating tumor DNA and RNA variants—ctDNA and ctRNA.
It is challenging to detect ct-nucleic acids (ctNA) in liquid biopsies due to their low concentrations and the low frequency of mutations when compared to the wild-type. Bioengineers have sought advances in nanoscience and nanotech to help address these challenges, which include scanning tunneling microscopy-assisted break junctions (STMBJ) that life scientists had recently used to detect the first single-molecule, by identifying RNA from E.coli strains.
This approach can be extended to detect cancer biomarker RNA sequences from human liquid biopsies. The method can detect small amounts of RNA biomarkers in solution via electrical means to obtain sequence-specific fingerprints that are sensitive to single-point mutations.
In this work, Gunasinghe et al. developed a proof-of-concept STMBJ method as a biosensor. The outcomes highlighted the first single molecule conductance results for sequences of human origin.
Mechanism of action
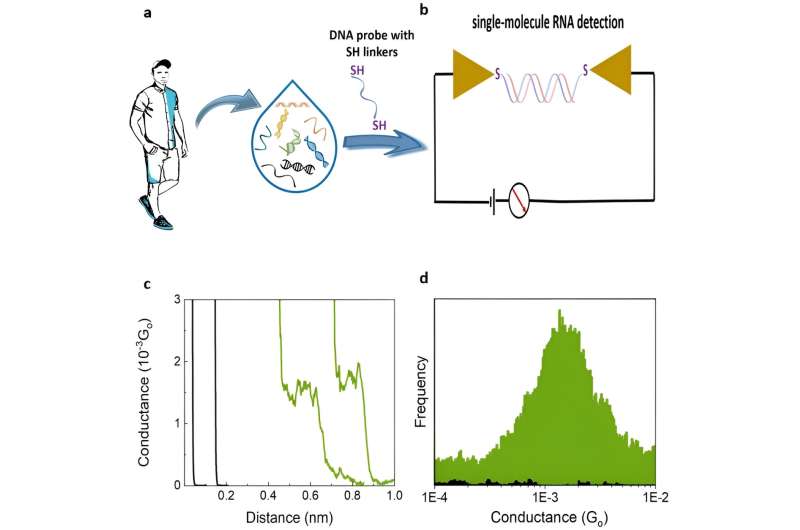
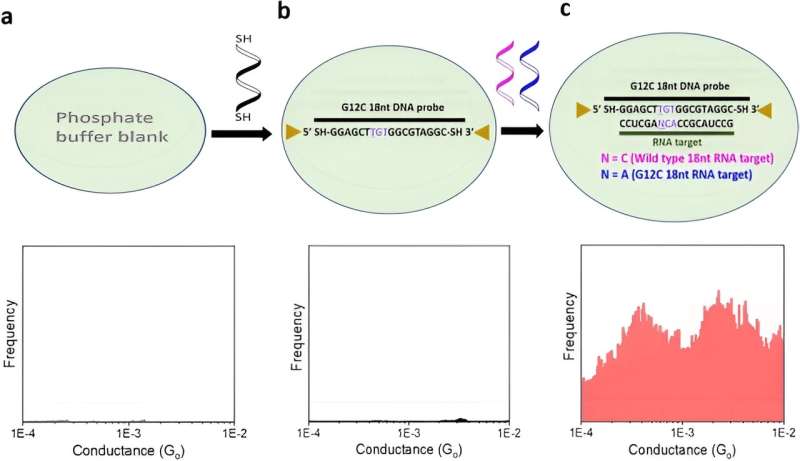
The team detected a liquid biopsy sample containing multiple ctDNA by targeting it with specific dithiol-modified DNA probes. They then electrically detected the single molecule RNA cancer biomarkers by hybridizing the DNA probe with the target RNA to create a DNA: RNA hybrid that bound to the scanning tunneling microscopy electrodes. The outcomes formed closed circuit biomolecular electronics to measure electrical fingerprints in the form of STMBJ conductance-distance curves to represent steps of molecular conductance.
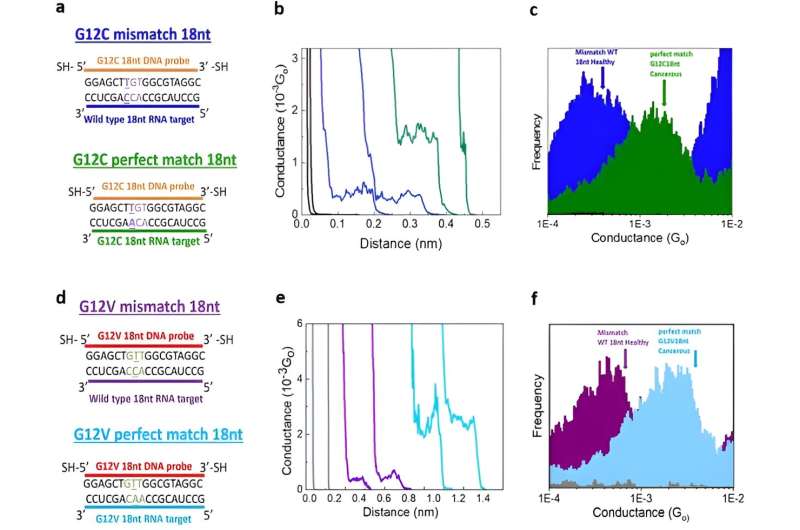
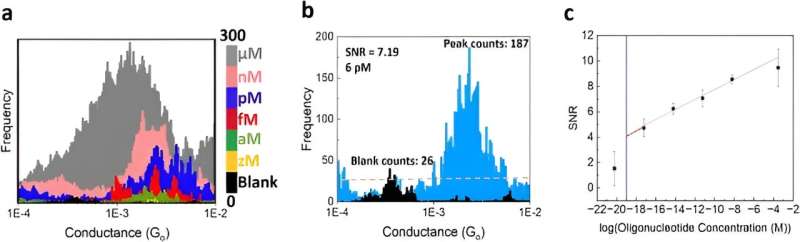
Gunasinghe and colleagues performed STMBJ experiments to electrically detect RNA cancer biomarkers. They measured the electrical conductance of two mutant variants and differentiated the mutation from the wild-type sequence simultaneously in solution. Using titration experiments, they showed a limit of detection with proof-of-concept electrical biosensors at the single molecule level with a high signal-to-noise ratio.
Designing biomolecular electronic DNA probes through bioinformatics
Genomics studies on cancer biomarkers include the Pancancer Analysis of Whole Genomes, the Consortium of the International Cancer Genome Consortium, and the Cancer Genome Atlas that has assessed whole cancer genomes from several tumor types.
This has created an opportunity to identify potential mutant sequences for RNA cancer biomarkers to be observed in liquid biopsy samples. The Kirsten rat sarcoma is a well-known oncogene with a high mutation rate in several cancers frequently found in pancreatic ductal adenocarcinoma, colorectal cancer and skin melanoma.
Such mutations typically contain a single base-level mutation occurring at codon 12 present in 46% of all lung adenocarcinomas known as G12, while this is present in a variety of colorectal and pancreatic cancers. The team focused on two specific mutations that covered a variety of cancer types with promising future screening approaches.
Electrical detection of single-molecule cancer biomarkers
Using the scanning tunneling microscopy break-junction method (STMBJ), Gunasinghe et al. measured the charge transport conductance values of individual mutant RNA molecules.
They recorded a range of conductance-distance curves. In the experimental setup, they modified the DNA probes with thiol binding groups to facilitate them linking to electrodes to hybridize with a complementary target RNA sequence and form a conductance step by closing the biomolecular electronics circuit. The sensitivity of the instrument prevented the observation of steps when there were no molecules bound between the two molecules.
This biosensing strategy allowed them to differentiate the mutants from the wild-type variants. The outcomes generated the first single-molecule measurements of an RNA sequence of human origin, while further elucidating it to be the first electrical detection of individual cancer biomarker molecules.
Specificity and sensitivity of the instrument
In this instance, Gunasinghe and colleagues defined specificity as the ability to discriminate cancer biomarkers from the wild-type sequence. The single-molecule electrical biosensor showed high specificity to detect the sequence of interest, allowing future applications to detect multiple disease-related sequences in parallel to diagnose a variety of diseases.
The team assessed the sensitivity of the approach through titration experiments to determine the limits of detection (LOD) of the system and defined the parameters of minimum target concentration. These outcomes provided a signal-to-noise ratio corresponding to the level of an individual molecule; a new target limit of detection. The titration experiments confirmed the superior sensitivity, and single-molecule nature of the biosensor to pave the way for cancer screening in a miniaturized, automated instrument in the future.
Outlook
In this way, Keshani G. Gunasinghe and colleagues presented a fully electrical proof-of-concept biosensor to detect RNA cancer biomarkers in solution with sensitivity at the level of a single molecule. The study represents a first application of single molecule measurements down to a sequence, for samples originating from humans.
Using the instrument, they efficiently identified the cancer biomarkers from the wild-type counterparts with a high signal-to-noise ratio. The extremely specific system simultaneously detected and differentiated multiple human RNA targets as healthy wild-type and mutant variants at a high specificity and sensitivity. The nanobiosensor has a promising future as an inexpensive, highly sensitive and label-free miniaturized device for early-stage cancer screening of liquid biopsies with future applications in biomedicine.
More information: Keshani G. Gunasinghe Pattiya Arachchillage et al, Electrical detection of RNA cancer biomarkers at the single-molecule level, Scientific Reports (2023). DOI: 10.1038/s41598-023-39450-6
Yuanhui Li et al, Detection and identification of genetic material via single-molecule conductance, Nature Nanotechnology (2018). DOI: 10.1038/s41565-018-0285-x
© 2023 Science X Network