This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New machine-learning method may aid personalized cancer therapy
Deep-learning technology developed by a team of Johns Hopkins engineers and cancer researchers can accurately predict cancer-related protein fragments that may trigger an immune system response. If validated in clinical trials, the technology could help scientists overcome a major hurdle to developing personalized immunotherapies and vaccines.
In a study published July 20 in the journal Nature Machine Intelligence, investigators from Johns Hopkins Biomedical Engineering, the Johns Hopkins Institute for Computational Medicine, the Johns Hopkins Kimmel Cancer Center and the Bloomberg~Kimmel Institute for Cancer Immunotherapy show that their deep-learning method, called BigMHC, can identify protein fragments on cancer cells that elicit a tumor cell-killing immune response, an essential step in understanding response to immunotherapy and in developing personalized cancer therapies.
"Cancer immunotherapy is designed to activate a patient's immune system to destroy cancer cells," says Rachel Karchin, Ph.D., professor of biomedical engineering, oncology and computer science, and a core member of the Institute for Computational Medicine. "A critical step in the process is immune system recognition of cancer cells through T cell binding to cancer-specific protein fragments on the cell surface."
The cancer protein fragments that elicit this tumor-killing immune response may originate from changes in the genetic makeup of cancer cells (or mutations), called mutation-associated neoantigens. Each patient's tumor has a unique set of such neoantigens that determine tumor foreignness, in other words, how different the tumor makeup is compared to self.
Scientists can identify which mutation-associated neoantigens a patient's tumor has by analyzing the genome of the cancer. Determining those which are most likely to trigger a tumor-killing immune response could enable scientists to develop personalized cancer vaccines or customized immune therapies as well as inform patient selection for these therapies. However, current methods for identifying and validating immune response-triggering neoantigens are time-consuming and costly, as these typically rely on labor-intense, wet laboratory experiments.
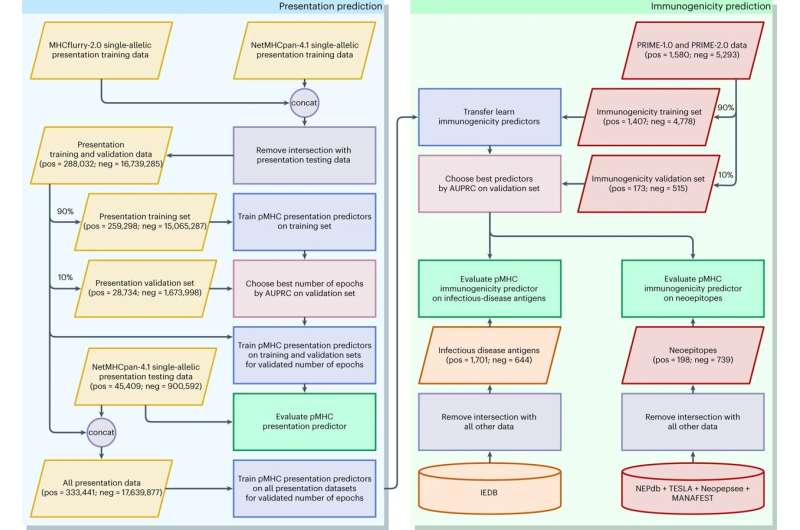
Because neoantigen validation is so resource intensive, there are few data to train deep-learning models. To address this, the researchers trained BigMHC, a set of deep neural networks, in a two-stage process called transfer learning. First, BigMHC learned to identify antigens that are presented at the cell surface, an early stage of the adaptive immune response for which many data are available.
Then, BigMHC was fine-tuned by learning a later stage, T-cell recognition, for which few data exist. In this manner, the researchers leveraged massive data to build a model of antigen presentation, and refined this model to predict immunogenic antigens.
The researchers tested BigMHC on a large independent data set, and showed that it was better at predicting antigen presentation than other methods. They further tested BigMHC on data from study co-author Kellie Smith, Ph.D., associate professor of oncology at the Bloomberg~Kimmel Institute for Cancer Immunotherapy, and found that BigMCH significantly outperformed seven other methods at identifying neoantigens that trigger T-cell response.
"BigMHC has outstanding precision at predicting immunogenic neoantigens," says Karchin.
"There is an urgent, unmet clinical need to tailor cancer immunotherapy to the subset of patients most likely to benefit, and BigMHC can shed light into cancer features that drive tumor foreignness, thus triggering an effective anti-tumor immune response," says study co-author Valsamo "Elsa" Anagnostou, M.D., Ph.D., director of the thoracic oncology biorepository, leader of the Johns Hopkins Molecular Tumor Board and Precision Oncology Analytics, and associate professor of oncology in the Kimmel Cancer Center.
The team is now expanding its efforts in testing BigMHC in several immunotherapy clinical trials to determine if it can help scientists sift through hundreds of thousands of neoantigens to filter down to those most likely to provoke an immune response.
"The hope is that BigMHC could guide cancer immunologists as they develop immunotherapies that can be used for multiple patients, or develop personalized vaccines that would boost a patient's immune response to kill their cancer cells," says lead author Benjamin Alexander Albert, who was an undergraduate student researcher in the departments of biomedical engineering and computer science at The Johns Hopkins University when the study was conducted. Albert is now a Ph.D. student at the University of California, San Diego.
Karchin and her team believe BigMHC and machine-learning-based tools like it can help clinicians and cancer researchers efficiently and cost-effectively sift through vast amounts of data needed to develop more personalized approaches to cancer treatment. "Deep learning has an important role to play in clinical cancer research and practice," Karchin says.
More information: Benjamin Alexander Albert et al, Deep neural networks predict class I major histocompatibility complex epitope presentation and transfer learn neoepitope immunogenicity, Nature Machine Intelligence (2023). DOI: 10.1038/s42256-023-00694-6