This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Unraveling the network of brain tumor genetics
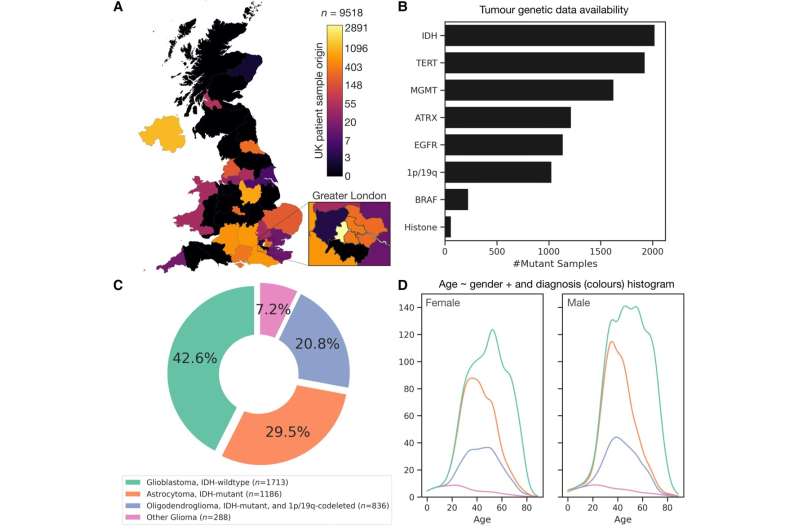
A new map of the network of genetic changes that drive the development of brain tumors has been revealed by UCL researchers. The genetic map, published in Brain, is comparable to a map of the London Underground—where different genetic features responsible for driving brain tumors are like stations connected by tracks.
Mapping the genetic changes should enable more accurate prediction of survival than standard diagnostic approaches.
Brain tumors are a common cause of death worldwide, with around 5,500 deaths due to brain tumors in the UK each year. However, the wide diversity of genetic changes that enable tumor cells to grow has made them remarkably difficult to treat.
Identifying effective treatments requires an understanding of the variable—and highly individual—relation between tumor genetics, patient outcomes, and disease mechanisms. Crucially, because this diversity is due to the interaction between multiple genetic changes in tumor development, it is ideally captured as a network.
Just as the shortest path from a given station to another is best found by analyzing the whole network of stations and tracks, the shortest path from normal to abnormal cells in an individual patient is best found by identifying the whole network of possible genetic changes leading to brain tumor development.
In the largest study of its kind, applying state-of-the-art network analysis to tumor genetic data from 4023 patients with brain tumors, drawn over 14 years across 12 countries, researchers at the UCL Queen Square Institute of Neurology have revealed a new map of the network of individual tumor genetic changes in patients with glioma, among the hardest brain tumors to treat.
Derived from data routinely collected during clinical care, the map is demonstrated to enable more accurate prediction of individual patient survival than the 'gold-standard' diagnostic labels adopted by the World Health Organization. Since the approach does not require new tests, it offers an efficient path to improved, more personalized care without any increase to health care costs.
Lead author, Dr. James Ruffle (UCL Queen Square Institute of Neurology), said, "The treatment of brain tumors is in desperate need of innovation: outcomes have hardly changed over the past 30 years. A critical step is gaining an understanding of tumor genetic diversity, which we show here to be accessible, but only to data and mathematical models of great size and complexity. A radical change of approach is needed, and we now have the computational tools to bring it into reality."
More information: James K Ruffle et al, Brain tumour genetic network signatures of survival, Brain (2023). DOI: 10.1093/brain/awad199