This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
3D genome architecture influences SCID-X1 gene therapy success
Patients with X-linked severe combined immunodeficiency disorder (SCID-X1), sometimes called "bubble boy disease," are born with a defective gene that prevents them from producing immune cells. Gene therapy from St. Jude Children's Research Hospital restored the immune system in multiple infants with SCID-X1 in 2019 by supplying copies of the corrected gene.
Through ongoing efforts to monitor patient safety, St. Jude scientists recently documented where the gene copies integrate into patient DNA, providing a foundation to understand the biology and safety of using lentiviral vectors. The findings were published today in Science Advances.
"We now have a robust pipeline to monitor the safety of lentiviral gene therapies," said senior co-corresponding author Jiyang Yu, Ph.D., St. Jude Department of Computational Biology interim chair. "This really gives hope to patients with genetic diseases that can be cured by lentiviral gene therapy. It looks like we cured bubble boy disease safely in these patients." The St. Jude team published the entire computational pipeline together with the manuscript.
Several years after treatment, the St. Jude lentiviral gene therapy for SCID-X1 appears both effective and safe, unlike earlier retroviral approaches. The researchers identified where the gene was added to patients' DNA and why the vector integrated in that particular place. Scientists previously knew that lentiviral gene therapies integrated into different areas of DNA that appeared safer than earlier technologies, but they could not address why.
3D genome structure holds the key to SCID-X1 gene therapy
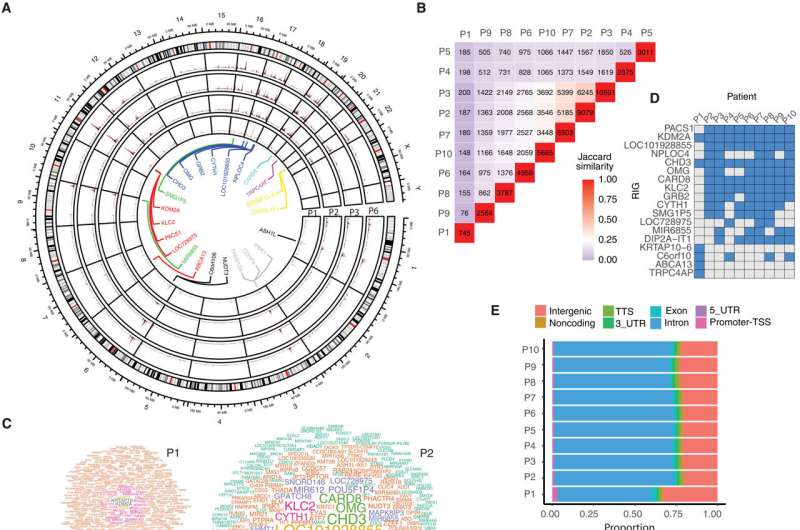
The scientists found that copies of the new, corrected gene had been inserted into certain genomic hotspots for the patients in the study. The reason was deceptively simple: From a 3D structural perspective, the hotspots are regions the lentiviral vector first encounters after entering the cell's nucleus through a channel called a nuclear pore.
"It's like someone coming into a room and taking the first available seat near the door," said co-corresponding Stephen Gottschalk, M.D., St. Jude Department of Bone Marrow Transplantation and Cellular Therapy chair. "The room is the nucleus. The seats are these DNA elements right near the door of the nuclear pore. That never occurred to me before this study, but it's a very simple principle in the end."
The St. Jude group found that the integration site pattern of the gene therapy into patient cells shed light on the safety and efficacy of the approach.
"We performed a comprehensive single-cell multi-omic analysis of this gene therapy to understand whether a functional copy of the corrected gene was in patient cells, to what extent the gene was expressed, and the chromatin organization at a single-cell level," said first author Koon-Kiu Yan, Ph.D., St. Jude Department of Computational Biology. "Before this study, we could measure the general gene expression of a bulk group of cells. But with bone marrow samples from two patients, we saw at a single-cell level which genes were expressed in which cell types."
Using that single-cell analysis, along with their other work, Yan showed that the safety and efficacy of treatment are also related to integration into compartments near the nuclear pore. Previous gene therapy efforts integrated into promoters near to or directly into oncogenes, ultimately causing cancer.
The lentiviral vector used by St. Jude selectively avoids promoters without disrupting oncogenes, improving treatment safety. The same pattern was observed with a lentiviral gene therapy used to create chimeric antigen receptor (CAR) T cells, suggesting that the phenomenon may be a general mechanism not restricted to the SCID-X1 vector.
"The integration pattern data could serve as a map of potentially safe integration sites," Yu and Gottschalk explained. "The single-cell analysis is like deep cartography, a map with a near pixel-perfect resolution. The large number of integration sites could be used as a safety reference for future lentiviral gene therapies."
One case was especially notable: the patient who required a second dose of the gene therapy to achieve a cure was found to have a different integration pattern than the patients who had responded to the first dose. After receiving the second dose, the patient's integration pattern resembled the others," and the therapy was effective. This study provides a foundation for understanding differences in treatment response so that future gene therapies may be improved.
More information: Koon-Kiu Yan et al, Integrome signatures of lentiviral gene therapy for SCID-X1 patients, Science Advances (2023). DOI: 10.1126/sciadv.adg9959. www.science.org/doi/10.1126/sciadv.adg9959