This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers discover unique differences in cancer cells based on where they are in a tumor
UCalgary researchers have discovered unique differences in cancer cells based on where they are in a tumor. The findings indicate the cells at the edge of a tumor may hold the key to guide treatment.
Taking a tumor biopsy is a critical first step used to gather essential information and diagnose disease. Generally, a biopsy is taken from the center of the suspicious tissue, mass, or lesion, however a recent University of Calgary study shows cells found at the edge of a tumor should be considered.
"Current technology allows us to look at tumor cells in a new way providing new understanding," says Dr. Pinaki Bose, Ph.D., assistant professor at the Cumming School of Medicine, and principal investigator. "We can see unique differences in cells found in the middle of a tumor compared with those found on the edge which can help guide treatment."
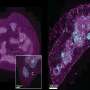
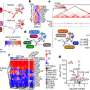
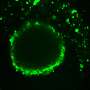
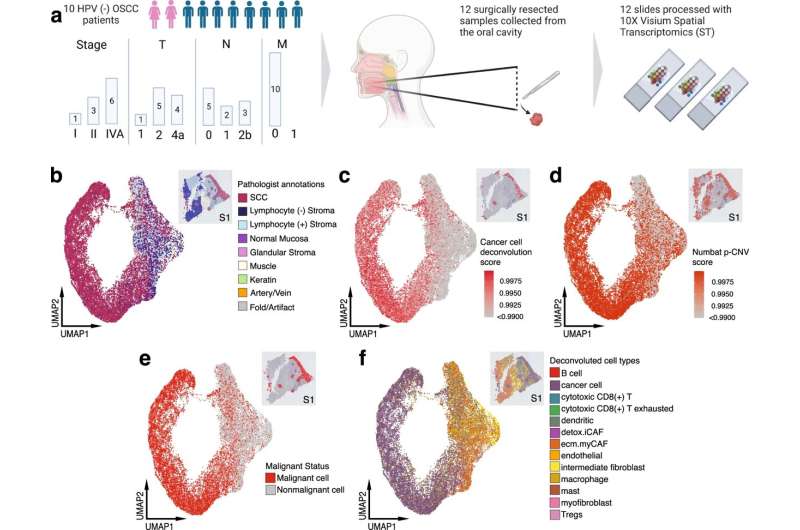
Examining the center of the tumor is generally the first step in discovery and allows for cancer staging, tumor grading, prognosis, and treatment plan. Using a technology called spatial transcriptomics, a molecular profiling method that preserves the positional identity of cells, the researchers were able to map and see important activity in a tumor including how the organization of cells affects the biology of a cancer.
The study published in Nature Communications showed important differences in the cells' architecture.
Analyzing a specific type of mouth cancer, oral squamous cell carcinoma, the researchers found that cells on the tumor's edge, where it interacts with normal tissue, provided very distinct information compared to cells from the middle. The team believes that this precise information could help in providing better understanding of the tumor, its aggressiveness, and the types of therapeutics it could respond best to.
"We also discovered that gene expression patterns in cells on the edge of a tumor aren't unique to mouth cancers," says Bose. "Our research showed similar results across several different cancer types, including breast, pancreatic, prostate and lung. This is an unexpected finding and could provide new targets for treatments."
Findings shows that the gene signature in cells at the edge of the tumor were associated with worse clinical outcomes across most cancer types, while the gene signature in cells at the core were associated with improved prognosis in cancers of similar origin to mouth cancers.
Bose says the use of spatial transcriptomics data and computational modeling paves the way for a deeper understanding of tumor biology and could unlock new information about how tumors invade healthy cells, what triggers metastasis and how these cells escape the body's immune response.
The study's co-first authors, Rohit Arora and Christian Cao were undergraduate students when the study was conducted, and Bose gives them a lot of credit for the important discoveries reported in the study. Both students continue their academic pursuit; Arora is a doctoral student at Harvard University and Cao is a medical student at the University of Toronto.
More information: Rohit Arora et al, Spatial transcriptomics reveals distinct and conserved tumor core and edge architectures that predict survival and targeted therapy response, Nature Communications (2023). DOI: 10.1038/s41467-023-40271-4