This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
AI accurately predicts cancer outcomes from tissue samples
Researchers at UT Southwestern Medical Center have developed a novel artificial intelligence (AI) model that analyzes the spatial arrangement of cells in tissue samples. This innovative approach, detailed in Nature Communications, has accurately predicted outcomes for cancer patients, marking a significant advancement in utilizing AI for cancer prognosis and personalized treatment strategies.
"Cell spatial organization is like a complex jigsaw puzzle where each cell serves as a unique piece, fitting together meticulously to form a cohesive tissue or organ structure. This research showcases the remarkable ability of AI to grasp these intricate spatial relationships among cells within tissues, extracting subtle information previously beyond human comprehension while predicting patient outcomes," said study leader Guanghua Xiao, Ph.D., Professor in the Peter O'Donnell Jr. School of Public Health, Biomedical Engineering, and the Lyda Hill Department of Bioinformatics at UT Southwestern. Dr. Xiao is a member of the Harold C. Simmons Comprehensive Cancer Center at UTSW.
Tissue samples are routinely collected from patients and placed on slides for interpretation by pathologists, who analyze them to make diagnoses. However, Dr. Xiao explained, this process is time-consuming, and interpretations can vary among pathologists. In addition, the human brain can miss subtle features present in pathology images that might provide important clues to a patient's condition.
Various AI models built in the past several years can perform some aspects of a pathologist's job, Dr. Xiao added—for example, identifying cell types or using cell proximity as a proxy for interactions between cells. However, these models don't successfully recapitulate more complex aspects of how pathologists interpret tissue images, such as discerning patterns in cell spatial organization and excluding extraneous "noise" in images that can muddle interpretations.
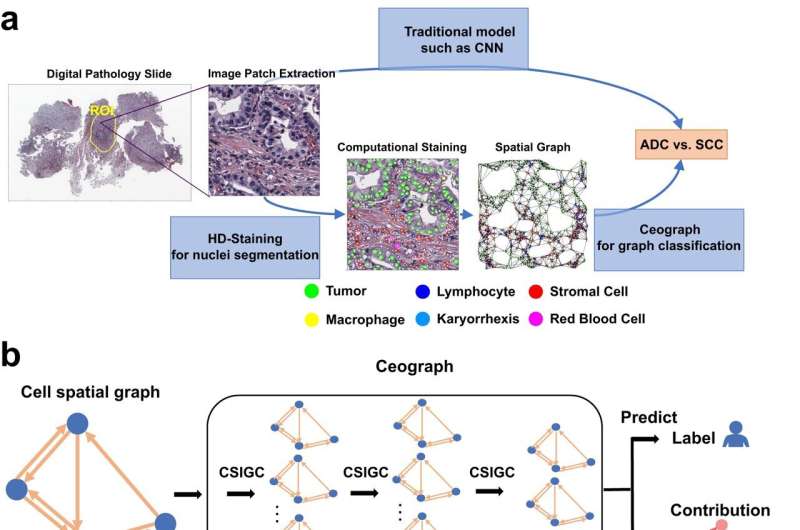
The new AI model, which Dr. Xiao and his colleagues named Ceograph, mimics how pathologists read tissue slides, starting with detecting cells in images and their positions. From there, it identifies cell types as well as their morphology and spatial distribution, creating a map in which the arrangement, distribution, and interactions of cells can be analyzed.
The researchers successfully applied this tool to three clinical scenarios using pathology slides. In one, they used Ceograph to distinguish between two subtypes of lung cancer, adenocarcinoma or squamous cell carcinoma. In another, they predicted the likelihood of potentially malignant oral disorders—precancerous lesions of the mouth—progressing to cancer. In the third, they identified which lung cancer patients were most likely to respond to a class of medications called epidermal growth factor receptor inhibitors.
In each scenario, the Ceograph model significantly outperformed traditional methods in predicting patient outcomes. Importantly, the cell spatial organization features identified by Ceograph are interpretable and lead to biological insights into how individual cell-cell spatial interaction change could produce diverse functional consequences, Dr. Xiao said.
These findings highlight a growing role for AI in medical care, he added, offering a way to improve the efficiency and accuracy of pathology analyses. "This method has the potential to streamline targeted preventive measures for high-risk populations and optimize treatment selection for individual patients," said Dr. Xiao, a member of the Quantitative Biomedical Research Center at UT Southwestern.
More information: Shidan Wang et al, Deep learning of cell spatial organizations identifies clinically relevant insights in tissue images, Nature Communications (2023). DOI: 10.1038/s41467-023-43172-8