This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
reputable news agency
proofread
What we know about autism—and how to treat it—could change after new UCSF study

By fusing the power of artificial intelligence with new molecular techniques that also seem ripped from science fiction, researchers at UCSF have mapped the microscopic world of autism spectrum disorder in unprecedented detail, pointing toward possible therapies for a subset of patients who have specific genetic mutations, according to a new study.
"This opens up kind of a Goldilocks of potential treatment targets," said one of the study's authors, Matthew State, a UCSF child psychiatrist and geneticist. "It's an opportunity for shots on goal that we just have not had before, because of the complexity of autism."
The results are novel for a few reasons, researchers said. It's the first time that the cellular workings of a neuropsychiatric disorder like ASD have been explored this deeply, opening a door for similar investigations of other neuropsychiatric disorders such as attention deficit hyperactivity disorder and schizophrenia.
The study also combined a basket of new technologies in an original way. Those technologies include stem cells, CRISPR-based genetic tools and AlphaFold 2, the Google AI that predicts the behavior of proteins.
In addition, researchers leveraged a system originally developed at UCSF to study viruses, including the pandemic-causing coronavirus. That platform is able to create comprehensive maps of the interactions between proteins, the biological machines produced by genes that carry out work in the cell.
"We've been at the bleeding edge of putting all this together," said another study leader, Nevan Krogan, director of the Quantitative Biosciences Institute within UCSF's School of Pharmacy, which coordinated the research along with the Department of Psychiatry and Behavioral Sciences.
The paper was posted Monday on bioRxiv.org, a preprint server, and will be submitted to a peer-reviewed journal. UCSF's Jeremy Willsey and Tomasz Nowakowski co-led the work, and Rezo Therapeutics, a biotech company founded by QBI and Krogan, also contributed.
Over the last decade, scientists including State have discovered over 100 mutated genes that are connected to autism. But translating that knowledge into therapies has been difficult, State said. The brain is intricate, and just knowing the right genes isn't enough.
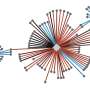
The UCSF-led team took a new approach, focusing on the proteins manufactured by those genes and mapping how they interact with each other.
Researchers found 1,000 proteins and more than 1,800 interactions. About 90% of the interactions "are things we've never seen before," State said.
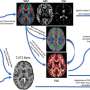
The team then looked for clues about how the mutations lead to the disorder by using specific mutations from a subset of ASD patients. The scientists exploited AlphaFold's predictive abilities and various molecular techniques to pinpoint the most meaningful interactions, studying how those proteins function in human cells and in "brain organoids" grown from stem cells.
These findings could one day lead to new drugs for patients, State and Krogan said.
"We're shining a brand-new light on autism," Krogan said.
More information: Belinda Wang et al, A foundational atlas of autism protein interactions reveals molecular convergence, bioRxiv.org (2023). DOI: 10.1101/2023.12.03.569805
(c)2023 the San Francisco Chronicle
Distributed by Tribune Content Agency, LLC.