This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Mouse study uncovers how altered gene expression can induce autism
Autism spectrum disorder (ASD) encompasses neurodevelopmental conditions where patients display repetitive behavior and impaired sociality. Genetic factors have been shown to influence the development of ASD. Additionally, recent studies have shown that the genes involved in chromatin modification and gene transcription are involved in the pathogenesis of ASD.
Among the many genes implicated in this process, the gene KMT2C (lysine methyltransferase 2c), which codes for a catalytic unit of H3K4 (histone H3 lysine 4) methyltransferase complex, has been identified to be associated with the development of autism and other neurodevelopmental disorders.
Previous studies have shown that haploinsufficiency (a condition where, of the two copies of the gene, only one remains functional) of KMT2C is a risk factor for ASD and other neurodevelopmental disorders. However, the molecular mechanism through which the loss-of-function mutation in KMT2C leads to these conditions remains unclear.
To address this knowledge gap, researchers from Juntendo University, RIKEN, and the University of Tokyo in Japan aimed to provide answers to these questions in a benchmark study published in the journal Molecular Psychiatry on 26 March 2024. The research team included Professor Tadafumi Kato from the Department of Psychiatry and Behavioral Science at Juntendo University Graduate School of Medicine, Dr. Takumi Nakamura and Dr. Atsushi Takata from the RIKEN Center for Brain Science, and Professor Takashi Tsuboi from Graduate School of Arts and Sciences, The University of Tokyo.
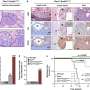
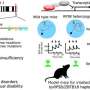
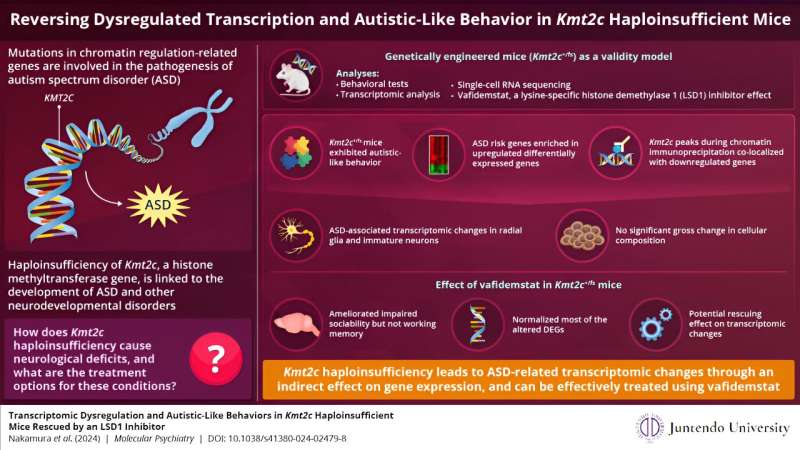
To get to the bottom of KMT2C's role in ASD pathogenesis, the team developed and analyzed genetically engineered strain mice (Kmt2c+/fs) having a frameshift mutation that models the KMT2C haploinsufficiency. They then performed various behavioral analyses, in which they observed that the mutant mice exhibited lower sociality, inflexibility, auditory hypersensitivity, and cognitive impairments, which are all ASD-related symptoms.
Next, they performed transcriptomic and epigenetic profiling to understand the basis of the molecular changes observed in the mutant mice. What they discovered was remarkable: the genes associated with increased ASD risk showed higher expression in these mutant mice. Dr. Takata says, "This was somewhat unexpected. KMT2C mediates H3K4 methylation, which is thought to activate gene expression, and thereby KMT2C haploinsufficiency was expected to cause reduced expression of target genes."
To gain mechanistic insights into their finding, the researchers carried out chromatin immunoprecipitation, a technique to determine the location on the DNA where the protein interacts with it. They found an overlap between KMT2C and the differentially expressed genes exhibiting reduced expression, suggesting that KMT2C haploinsufficiency leads to ASD-related transcriptomic changes through an indirect effect on gene expression.
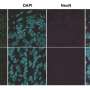
To identify the cell types that contribute more to the pathological changes seen in the mutant mice, the researchers performed single-cell RNA sequencing of newborn mice brains. They observed that the altered genes associated with ASD risk were predominant in undifferentiated radial glial cells. However, a gross change in the cell composition was not observed, implying that the transcriptomic dysregulation does not severely impact cell fate.
Finally, the researchers tested the effects of vafidemstat, a brain penetrant inhibitor of LSD1 (lysine-specific histone demethylase 1A), that could ameliorate histone methylation abnormalities. They found that vafidemstat improved the social deficits in the mutant mice and had an exceptional rescuing effect by changing the expression levels of the differentially expressed genes to their normal expression level. This finding showed that vafidemstat is a valid drug for mutant mice and can potentially help restore the normal transcriptomic state.
What sets this discovery apart is that it challenges the commonly held belief that ASD disability may not be cured and demonstrates the efficacy of vafidemstat in improving ASD-like phenotypes. The results open doors to future research to strengthen the foundation for the pharmacologic treatment of ASD and other neurodevelopmental disorders.
Prof. Kato concludes, "Our research shows that drugs similar to vafidemstat may be generalizable to multiple categories of psychiatric disorders."
More information: Takumi Nakamura et al, Transcriptomic dysregulation and autistic-like behaviors in Kmt2c haploinsufficient mice rescued by an LSD1 inhibitor, Molecular Psychiatry (2024). DOI: 10.1038/s41380-024-02479-8