This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Bridging diet, microbes, and metabolism: Implications for metabolic disorders
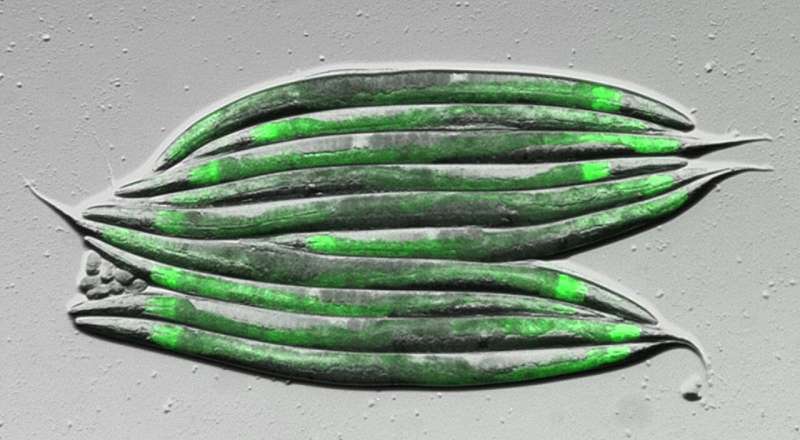
Mounting evidence suggests that the secret to understanding human health and combating metabolic diseases lies hidden within the microscopic world of our gut bacteria.
Recent research by scientists at the Boyce Thompson Institute (BTI) and Cornell University reveals that a specific fatty acid produced by gut bacteria directly influences fat metabolism in animals. This research is pivotal as it sheds light on the complex interplay between the diet, gut microbiota, and host metabolic health, offering insights that could open new avenues in our approach to managing metabolic disorders.
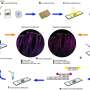
The researchers focused on certain gut bacteria that produce fatty acids with a special chemical structure known as a cyclopropane ring and showed that these can be converted into signals that turn on fat desaturation in the nematode C. elegans, a model organism often used to study human biology. Intriguingly, C. elegans itself produces a chemically similar fatty acid compound that regulates the same metabolic pathway as the bacterial cyclopropane fats.
"Our research suggests that the host organism may have acquired the ability to produce its own signaling molecule, mimicking bacterial biochemistry, through a gene obtained from bacteria—a process known as horizontal gene transfer," shared Bennett Fox, a post-doctoral researcher at BTI and first author of the study.
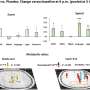
The research, recently published in Nature Communications, showed that both the bacterial and endogenous fatty acids activate a host receptor that functions as a central regulator of overall fat metabolism. This direct link between microbiota metabolites and host lipid metabolism offers insight into how our bodies could harness beneficial gut bacteria to regulate vital processes like obesity and metabolic dysfunction.
"Microbiota-dependent metabolites regulate virtually every aspect of animal physiology, including development, metabolism, and immune responses. Despite the life-sustaining importance of these metabolites, many of their structures remain unknown," noted Frank Schroeder, a professor at BTI and senior author of the study.
The fact that chemicals produced by bacteria can influence the metabolism of their host is a promising area of research. Further studies can investigate host-bacterial interactions to understand better—and potentially improve—metabolic health.
"The devil is in the details. As we gain clarity regarding the molecular mechanisms of fat metabolism and its regulation by specific diet-derived compounds, we step closer to harnessing this knowledge for better health outcomes in humans," said Fox. "This research not only broadens our understanding of basic biological processes but also highlights potential pathways for future exploration in human health and disease management."
More information: Bennett W. Fox et al, Evolutionarily related host and microbial pathways regulate fat desaturation in C. elegans, Nature Communications (2024). DOI: 10.1038/s41467-024-45782-2