This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Proteogenomics reveal prognostic markers of small cell lung cancer, advance development of precision therapies
Researchers in China have reported the first large-scale study characterizing the proteomics and phosphoproteomics of small cell lung cancer (SCLC) clinical cohorts, providing a comprehensive picture of the proteogenomics landscape of SCLC.
The team is led by Prof. Zhang Peng from Shanghai Pulmonary Hospital of the Tongji University, Prof. Zhou Hu from Shanghai Institute of Materia Medica of the Chinese Academy of Sciences (CAS), and Prof. Gao Daming and Prof. Ji Hongbin from the Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology of CAS.
This study, published in Cell, reveals the molecular features of SCLC and proposed new molecular subtypes and targeted personalized treatment strategies, and laying a solid foundation for a better understanding of the underlying mechanisms and improvement of clinical therapeutic strategies for SCLC.
Lung cancer is the most common malignant tumor worldwide. SCLC accounts for about 15% of all lung cancer cases, and it is characterized by rapid proliferation, strong metastatic potential, high treatment resistance, and the highest malignancy and fatality rate with a five-year survival rate of only 5%. However, the progress in the treatment of SCLC has been slow, and viable therapeutic targets and precision therapies that significantly improve survival rates have yet to be identified.
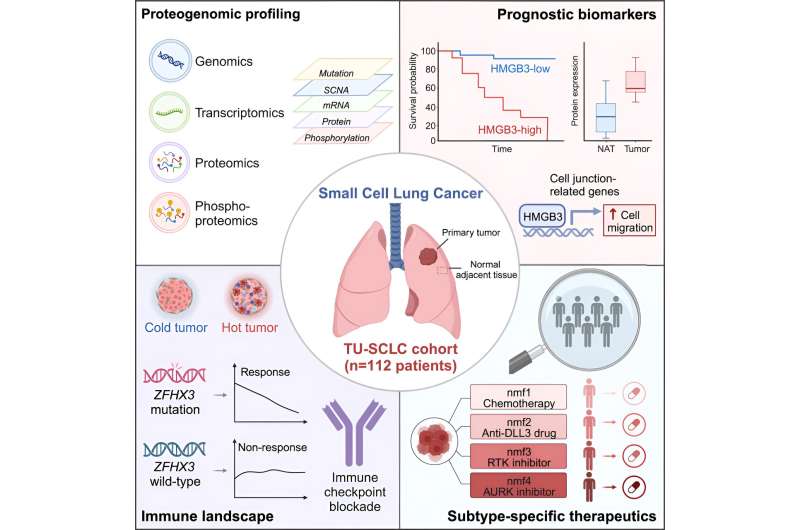
In this study, the researchers collected tumor and matched adjacent tissue samples from 112 Chinese SCLC patients who underwent surgical resection, and conducted integrative multi-omics analyses including whole exome sequencing, transcriptomics, proteomics, and phosphoproteomics.
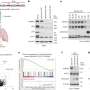
The researchers first identified TP53 and RB1 as the major gene alterations in SCLC through somatic mutations and copy number losses. It was found that the gene mutation frequency of ZFHX3 was higher in the Chinese populations compared to Western populations. The gene mutations of TP53, FAT1 and GNAS, were shown to have an impact on their own expressions, and FAT1 gene mutations could promote the occurrence and development of SCLC by promoting cell EMT phenotype.
Copy number variations were shown to have cis and trans regulatory effects on mRNA and protein expression, with RB1 loss showing a significant trans effect on the protein level but a minor effect on mRNA. Chromosome 5q loss regulated the expression of proteins related to DNA synthesis, DNA damage repair, and cell cycle through trans effects, which may be an important mechanism underlying SCLC development.
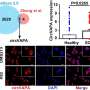
By comparing the proteomic characteristics of tumor tissues and matched adjacent tissues, the researchers then identified a series of proteins, phosphorylation sites and kinases (CHEK1, ATR, ATM, CDK2, GSK3A) that are closely associated with the occurrence and development of SCLC. HMGB3 and CASP10 were identified as significantly associated with prognosis. HMGB3 was highly expressed in tumors and was associated with worse prognosis. CASP10 had lower expression and was also correlated with poorer prognosis.
Immunohistochemical analysis in an independent clinical cohort confirmed the correlation between HMGB3 and CASP10 expression and prognosis, suggesting their potential value as prognostic indicators. Further biological experiments showed that HMGB3 could promote cell migration in SCLC through transcriptional regulation of cell junction-related gene expression.
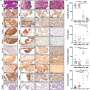
Besides, the researchers immunotyped SCLC tumors based on the levels of immune cell infiltration. Most SCLC tumors were classified as cold tumors with poor prognosis. Only a minority belonged to the hot tumor subtype, which showed activation of immune-related pathways such as interferon-gamma pathway, antigen presentation, and upregulation of immune checkpoint molecules (such as CTLA4, PD-L1, PD-1), suggesting potential benefits from immunotherapy.
ZFHX3 mutations were significantly enriched in the hot tumor subtype and were closely associated with increased immune cell infiltration. Patients with ZFHX3 mutations showed better treatment response, indicating that ZFHX3 mutations could be a biomarker for immunotherapy in SCLC and providing important clues for more precise tumor immunotherapy.
Finally, 107 SCLC tumor samples were divided into four distinct subtypes (nmf1~nmf4). Each subtype showed specific treatment vulnerabilities. For example, the nmf2 subtype showed high expression of DLL3, suggesting a potential better response to anti-DLL3 therapies like Tarlatamab, a bispecific antibody. The nmf3 subtype exhibited characteristics of elevated RTK signaling, indicating potential benefits from treatments with RTK inhibitors such as Anlotinib. The discovery of these subtypes and their corresponding features will aid in guiding the clinical treatment selection for SCLC.
The researchers also established large-scale patient-derived tumor xenograft (PDX) models and cell line-derived xenograft (CDX) models of SCLC, and conducted genomic and proteomic integration analysis on them to validate the targeted treatment strategies based on the molecular subtyping of clinical samples.
In summary, these findings provide a theoretical basis for the understanding of the pathogenic mechanisms, prognosis prediction, molecular subtyping, and personalized treatment of SCLC. The high-quality data generated will support both basic and clinical research in the field of SCLC.
More information: Qian Liu et al, Proteogenomic characterization of small cell lung cancer identifies biological insights and subtype-specific therapeutic strategies, Cell (2024). DOI: 10.1016/j.cell.2023.12.004