This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Team develops a single-nucleus resolution atlas of white adipose tissue in different depots
Adipose tissue is an important regulator of metabolism and energy homeostasis in the human body. It is usually classified into two distinct categories: white adipose tissue (WAT) and brown adipose tissue (BAT). WAT is widely distributed throughout the body of mammals, and based on their anatomical distribution, can be divided into subcutaneous adipose tissue (SAT), visceral adipose tissue (VAT), and bone marrow adipose tissue (BMAT).
Energy storage is a conserved function of adipose tissue, but in recent years, researchers have found that WAT in different adipose depots exhibits unique biological characteristics and has specific endocrine functions, leading to a growing interest in adipose tissue heterogeneity.
A number of studies have examined cellular heterogeneity in adipose tissue by using single-cell RNA sequencing (scRNA-seq). However, due to the limitations of scRNA-seq in sequencing large-size adipocytes, these studies have mainly focused on the heterogeneity of the stromal vascular fraction (SVF) or the identification of adipose stem cells, new subpopulations of immune cells, and disease-specific subpopulations within adipose tissues, and little research has been done on the heterogeneity of adipose tissues themselves under physiologic and pathologic conditions.
With the development of single-nucleus RNA sequencing (snRNA-seq) technology that has successfully overcome the limitations of conventional scRNA-seq analysis for large adipocytes, we can now understand the heterogeneity of white and brown adipocytes as well as their interaction with stromal vascular cells. Nevertheless, previous studies utilizing snRNA-seq have often focused on one or two adipose depots and could not fully answer the question of heterogeneity of adipose depots in different anatomical sites.
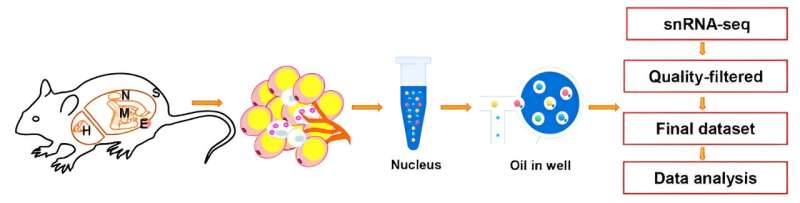
Recently, Tuo Deng's lab at The Second Xiangya Hospital of Central South University published a study in Life Metabolism titled "Single-nucleus RNA sequencing reveals heterogeneity among multiple white adipose tissue depots." Using snRNA-seq technology, the researchers examined five adipose depots, including subcutaneous (SWAT), epididymal (EWAT), mesenteric (MWAT), perirenal (NWAT), and pericardial (HWAT) tissues under physiological conditions in mice, and generated cellular maps of multiple adipose depots, as well as analyzing the heterogeneity among the depots in terms of cellular composition, interactions, and transcriptional profiles.
These data revealed a diversity of cell types including adipocytes (AD), immune cells (IC), endothelial cells (EC), mesothelial cells (MC), fibroblast-adipogenic progenitors (FAP), and pericytes (PC). Further analysis of the cellular composition showed that the heterogeneity of ICs, MCs, and FAPs was particularly pronounced, while the heterogeneity of adipocytes and ECs was relatively small. The differences in primitive IC subpopulations in WATs at different sites were also analyzed by flow cytometry and RNA-seq, and were experimentally verified by hematoxylin and eosin (H&E) staining.
The subsequent in-depth analysis of the sequencing data revealed that approximately 30% of the total adipose tissue cell population were MCs and ECs, providing a new theoretical basis for the anatomical basis of obesity-induced lymphatic vessel growth and dysfunction in MWAT.
In conclusion, the study explored the heterogeneity of adipose tissue under physiological conditions from a new perspective by analyzing adipose tissue from five depots using snRNA-seq. The results contribute to a deeper understanding of adipose tissue biology and open avenues for further research on the role of adipose tissue in health and disease.
More information: Limin Xie et al, Single-nucleus RNA sequencing reveals heterogeneity among multiple white adipose tissue depots, Life Metabolism (2023). DOI: 10.1093/lifemeta/load045