This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
'Virtual biopsy' uses AI to help doctors assess lung cancer
Imperial researchers have used artificial intelligence (AI) to extract information about the chemical makeup of lung tumors from medical scans. For the first time, they have demonstrated how combining medical imaging with AI can be used to provide a 'virtual biopsy' for cancer patients.
Their non-invasive method can classify the type of lung cancer a patient has—which is crucial in selecting the right treatment—and can predict if the cancer is likely to progress. According to the researchers, the technique could be used by doctors when it's not possible or suitable to obtain a physical tissue biopsy from a patient.
The study is published in the journal npj Precision Oncology and was led by Imperial College London, alongside collaborators in Córdoba, Spain.
The study's senior author, Professor Eric Aboagye, from Imperial's Department of Surgery and Cancer, said, "At the moment, trying to find out in-depth information about tissues and tumors requires invasive biopsies, which can be uncomfortable for the patient, delay treatment decisions, and be costly for health services. Although CT scans are commonly used in the clinic, they fall short of offering detailed insights into the cellular type or prognostic information of diseases."
First-author and Imperial Ph.D. candidate Marc Boubnovski Martell adds, "We've developed a system that merges CT scans with the chemical makeup of tumors and normal lung tissue. This allows us to classify lung cancer types and, importantly, provides reliable predictions about patient outcomes."
Early detection and diagnosis
Lung cancer is the most common cause of cancer death in the UK, with around 35,000 lives lost to the disease each year, according to Cancer Research UK. This is partly because symptoms don't appear in the early stages, and there is a pressing need for new ways to detect and treat the tumor before it spreads to other parts of the body.
Patients who present with symptoms of lung cancer tend to be diagnosed using chest X-rays and computed tomography (CT) scans—which can also show if cancer has spread beyond the lungs. If it's safe to get a biopsy sample, clinical scientists then look at the tumor cells under a microscope and classify the type of lung cancer a patient has. This helps doctors decide which course of treatment might be best.
A relatively new test called metabolomic profiling, which also requires a tissue biopsy, can provide much more detailed information about tumor cell chemistry and metabolism and, importantly, how the cancer is likely to evolve. However, it is labor- and time-intensive and so isn't performed routinely in hospitals.
AI-powered imaging
In recent years, AI has been used to analyze medical scans and look for signs of disease that can be missed by doctors or might not even be visible to the naked eye. Generative AI, a type of AI that is able to learn from data to create new content, is currently being pursued for multiple applications.
The Imperial team took these ideas a step further and wondered if information about lung tumor chemistry contained in the metabolomic profile might show up in CT scans.
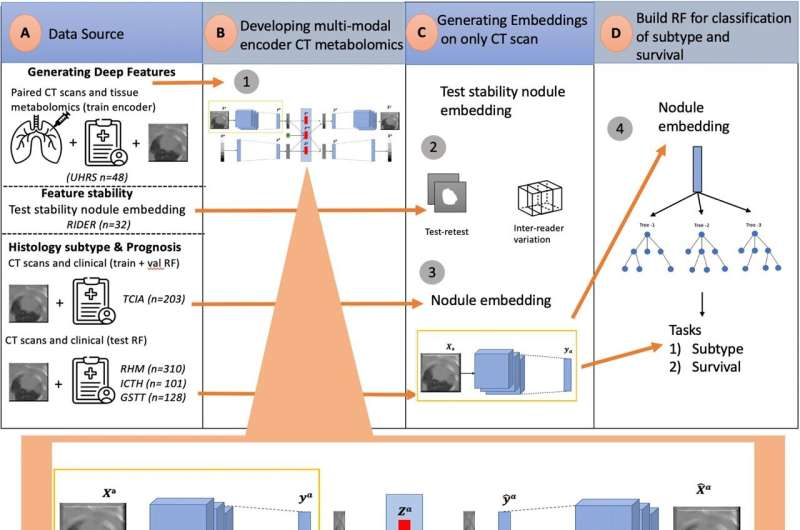
First, though, any AI model needs to be trained on existing groups of patients who have a medical scan, a definitive diagnosis, and preferably lots of additional clinical information. The researchers used data from 48 lung cancer patients who were treated at the University Hospital Reina Sofia (UHRS) in Córdoba, Spain.
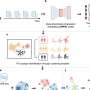
Uniquely, all the patients had a CT scan performed, as well as detailed metabolomic profiling of their tumor tissue and healthy tissue next to the tumor. Based on this data, the Imperial team developed an AI-powered, deep-learning assessment tool they called tissue-metabolomic-radiomic-CT (TMR-CT).
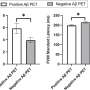
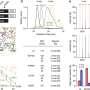
The researchers found a significant and powerful correlation between patients' metabolomic profiles and the 'deep features' of their CT scans, which appear as brighter or darker areas in the image.
Using this method, the researchers theorized they could bypass the need for physical tissue samples and infer tumor metabolic characteristics from the CT scan alone.
To test this, they used their TMR-CT model in a separate group of 723 lung cancer patients who were treated at Royal Marsden Hospital, Guy's and St Thomas' Hospital, or Imperial College NHS Healthcare Trust. All patients had a CT scan, but there was no available metabolomics data.
The results showed that TMR-CT adeptly classified lung cancer and, importantly, gave dependable predictions about patient outcomes, surpassing the performance of traditional CT-based methods and clinical assessments.
A future clinical tool
The researchers hope to confirm their TMR-CT method in other groups of lung cancer patients as well as potentially people with brain, ovarian, and endometrial cancers, which can also be difficult to obtain biopsies.
In the future, the technique could be incorporated as an algorithm as part of the software loaded onto commercial medical imaging scanners.
Professor Aboagye concludes, "This research shows the potential of using CT scans to gain a deeper, more nuanced understanding of tissue and tumor chemical composition, that has until now only been accessible through direct tissue sampling. This method could prove particularly beneficial in countries like the UK, where lung cancer prevalence is high, and potentially transform diagnostic and treatment protocols."
More information: Marc Boubnovski Martell et al, Deep representation learning of tissue metabolome and computed tomography annotates NSCLC classification and prognosis, npj Precision Oncology (2024). DOI: 10.1038/s41698-024-00502-3