This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Genome-wide transcriptome profiling and development of age prediction models in the human brain
A new research paper was published in Aging entitled "Genome-wide transcriptome profiling and development of age prediction models in the human brain."
Aging-related transcriptome changes in various regions of the healthy human brain have been explored in previous works. However, a study to develop prediction models for age based on the expression levels of specific panels of transcripts is lacking.
Moreover, studies that have assessed sexually dimorphic gene activities in the aging brain have reported discrepant results, suggesting that additional studies would be advantageous. The prefrontal cortex (PFC) region was previously shown to have a particularly large number of significant transcriptome alterations during healthy aging in a study that compared different regions in the human brain.
In this new study, researchers Joseph A. Zarrella and Amy Tsurumi from the Harvard T.H. Chan School of Public Health, Massachusetts General Hospital, Harvard Medical School, and Shriner's Hospitals for Children-Boston aimed to profile PFC transcriptome changes during healthy human aging overall and comparing potential differences between female and male samples, as well as developing chronological age prediction models by various methods.
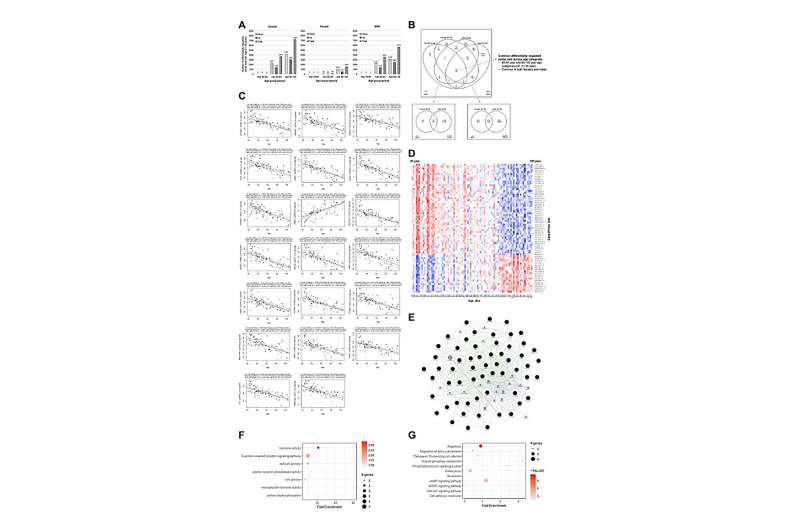
"We harmonized neuropathologically normal PFC transcriptome datasets obtained from the Gene Expression Omnibus (GEO) repository, ranging in age from 21 to 105 years, and found a large number of differentially regulated transcripts in the old and elderly, compared to young samples overall, and compared female and male-specific expression alterations."
The team assessed the genes that were associated with age by employing ontology, pathway, and network analyses. Furthermore, they applied various established (least absolute shrinkage and selection operator (Lasso) and Elastic Net (EN)) and recent (eXtreme Gradient Boosting (XGBoost) and Light Gradient Boosting Machine (LightGBM)) machine learning algorithms to develop accurate prediction models for chronological age and validated them.
Studies to further validate these models in other large populations and molecular studies to elucidate the potential mechanisms by which the transcripts identified may be related to aging phenotypes would be advantageous.
"Our results support the notions that specific gene expression changes in the PFC are highly correlated with age, that some transcripts show female and male-specific differences, and that machine learning algorithms are useful tools for developing prediction models for age based on transcriptome information."
More information: Joseph A. Zarrella et al, Genome-wide transcriptome profiling and development of age prediction models in the human brain, Aging (2024). DOI: 10.18632/aging.205609