This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
A computational workflow that predicts metabolites and metabolic pathways associated with somatic mutations in cancers
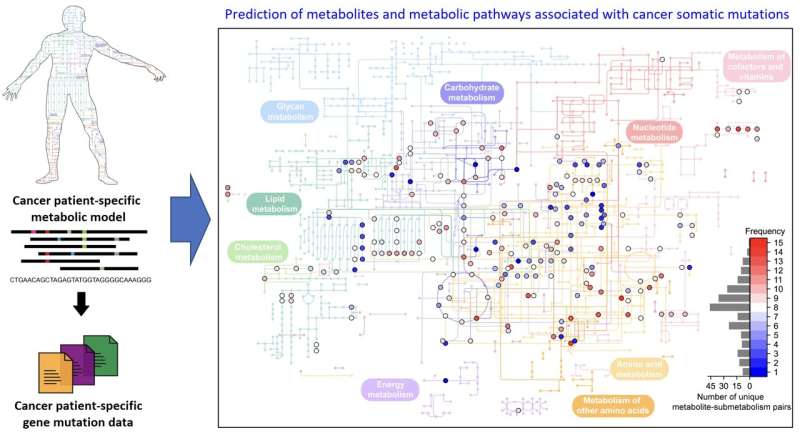
Cancer is characterized by abnormal metabolic processes different from those of normal cells. Therefore, cancer metabolism has been extensively studied to develop effective diagnosis and treatment strategies.
Notable achievements of cancer metabolism studies include the discovery of oncometabolites and the approval of anticancer drugs by the U.S. Food and Drug Administration (FDA) that target enzymes associated with oncometabolites. An oncometabolite is a metabolite that shows pro-oncogenic function when abnormally accumulated in cancer cells. An oncometabolite is often generated as a result of gene mutations, and this accumulation promotes the growth and survival of cancer cells. Representative oncometabolites include 2-hydroxyglutarate, succinate, and fumarate.
Approved anticancer drugs such as "Tibsovo (active ingredient: ivosidenib)" and "Idhifa (active ingredient: enasidenib)" are both used for the treatment of acute myeloid leukemia. Despite such achievements, studying cancer metabolism, especially oncometabolites, remains challenging due to time-consuming and expensive methodologies such as metabolomics. Thus, the number of confirmed oncometabolites is very small although a relatively large number of cancer-associated gene mutations have been well studied.
On March 18, a KAIST research team led by Professor Hyun Uk Kim from the Department of Chemical and Biomolecular Engineering developed a computational workflow that systematically predicts metabolites and metabolic pathways associated with somatic mutations in cancer through collaboration with research teams under Prof Youngil Koh, Prof. Hongseok Yun, and Prof. Chang Wook Jeong from Seoul National University Hospital.
The research is published in the journal Genome Biology in a paper titled "Prediction of metabolites associated with somatic mutations in cancers by using genome‑scale metabolic models and mutation data."
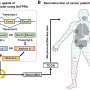
The research teams have successfully reconstructed patient-specific genome-scale metabolic models (GEMs; a computational model that mathematically describes all of the biochemical reactions that take place inside a cell) for 1,043 cancer patients across 24 cancer types by integrating publicly available cancer patients' transcriptome data (i.e., from international cancer genome consortiums such as PCAWG and TCGA) into a generic human GEM. The resulting patient-specific GEMs make it possible to predict each patient's metabolic phenotypes.
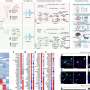
The team developed a four-step computational workflow using the patient-specific GEMs from 1,043 cancer patients and somatic mutation data obtained from the corresponding cancer patients. This workflow begins with the calculation of the flux-sum value of each metabolite by simulating the patient-specific GEMs.
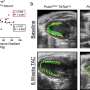
The flux-sum value quantifies the intracellular importance of a metabolite. Next, the workflow identifies metabolites that appear to be significantly associated with specific gene mutations through a statistical analysis of the predicted flux-sum data and the mutation data. Finally, the workflow selects altered metabolic pathways that significantly contribute to the biosynthesis of the predicted oncometabolite candidates, ultimately generating metabolite-gene-pathway sets as an output.
The two co-first authors, Dr. GaRyoung Lee (currently a postdoctoral fellow at the Dana-Farber Cancer Institute and Harvard Medical School) and Dr. Sang Mi Lee (currently a postdoctoral fellow at Harvard Medical School) said, "The computational workflow developed can systematically predict how genetic mutations affect cellular metabolism through metabolic pathways. Importantly, it can easily be applied to different types of cancer based on the mutation and transcriptome data of cancer patient cohorts."
Prof. Kim said, "The computational workflow and its resulting prediction outcomes will serve as the groundwork for identifying novel oncometabolites and for facilitating the development of various treatment and diagnosis strategies."
More information: GaRyoung Lee et al, Prediction of metabolites associated with somatic mutations in cancers by using genome-scale metabolic models and mutation data, Genome Biology (2024). DOI: 10.1186/s13059-024-03208-8