This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Researchers unveil faster and more accurate COVID test
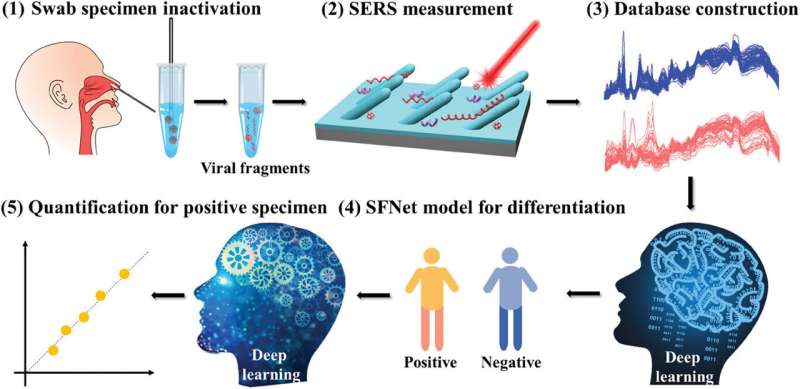
With new cases, hospitalizations and mortality rates holding steady in many parts of the world, University of Georgia researchers have developed a faster detection technique for COVID-19.
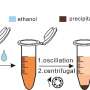
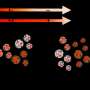
In a new study published in Advanced Materials Interface, the UGA research team describes the rapid diagnostic test for SARS-CoV-2 detection and quantification directly from human nasopharyngeal swabs using surface-enhanced Raman spectroscopy and deep learning algorithms.
The overall process for the test is comparable to the results from polymerase chain reaction (PCR), for COVID-19 and other respiratory virus infection diagnosis tests, using a simple procedure requiring less than 15 minutes.
"The new technique has several advantages over current methods, including speed, accuracy, and the quantification of viral load beyond simply 'positive or negative,'" said Yiping Zhao, Distinguished Research Professor in the Franklin College of Arts and Sciences department of physics and astronomy.
"This method is easy to adapt to multiplex detection, able to be generalized to predict both virus variant species and concentrations simultaneously for a panel of viruses."
Unlike the PCR test, which requires specific labels and probes for target amplification and detection, the SERS-based method does not require any labels. According to the researchers, this simplifies the diagnostic process, reduces costs, and eliminates the risk of false positives or negatives due to labeling errors.
The SERS-based capability to quantify the viral load for positives directly from clinical specimens is valuable for monitoring disease progression, assessing treatment efficacy, and making informed clinical decisions.
"The methodology is validated using actual clinical specimens, demonstrating its effectiveness in real-world scenarios," said Yanjun Yang, UGA post-doctoral researcher and co-author who recently interviewed more than 40 health care professionals about the use of rapid diagnostics. "This real-world applicability is essential for point-of-care diagnostics, especially in resource-limited settings or during outbreaks."
Currently, the UGA team's method is designed for diagnostic laboratories to facilitate large-scale screenings for patient infections, as indicated by the price of instruments. While COVID-19 self-test kits offer convenience and accessibility, they also come with several disadvantages that could eventually be replaced by Raman instruments.
"If the price becomes affordable for individuals through large-scale production, a 'car key' size Raman instrument would facilitate widespread adoption," Yang said. "Furthermore, if this device integrates with our database, which contains a variety of virus SERS spectra including a panel of respiratory viruses (COVID-19, Flu, RSV, etc.) as well as their mutations or variants, it could become a powerful self-testing tool."
More information: Yanjun Yang et al, Advancing SERS Diagnostics in COVID‐19 with Rapid, Accurate, and Label‐Free Viral Load Monitoring in Clinical Specimens via SFNet Enhancement, Advanced Materials Interfaces (2024). DOI: 10.1002/admi.202400013