This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Propensity score matching offers more efficient biomarker discovery in cancer research
Thanks to advances in molecular sequencing, scientists have generated a wealth of data from the genome, transcriptome, proteome and metabolome. However, these "-omics" datasets are complex and there is currently no standard method for parsing through the data to separate specific biomarkers from possibly confounding variables.
In a recent study, scientists at Beth Israel Deaconess Medical Center (BIDMC) evaluated a statistical technique already in use in other fields to minimize confounding factors for its efficiency in omics analysis in two colorectal (CRC) cancer data sets. The work is published in the journal PLOS ONE.
The team's findings suggest that the technique, known as propensity score matching (PSM), may emerge as an efficient and cost-effective strategy for multiomic data analysis and clinical trial design for biomarker discovery.
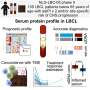
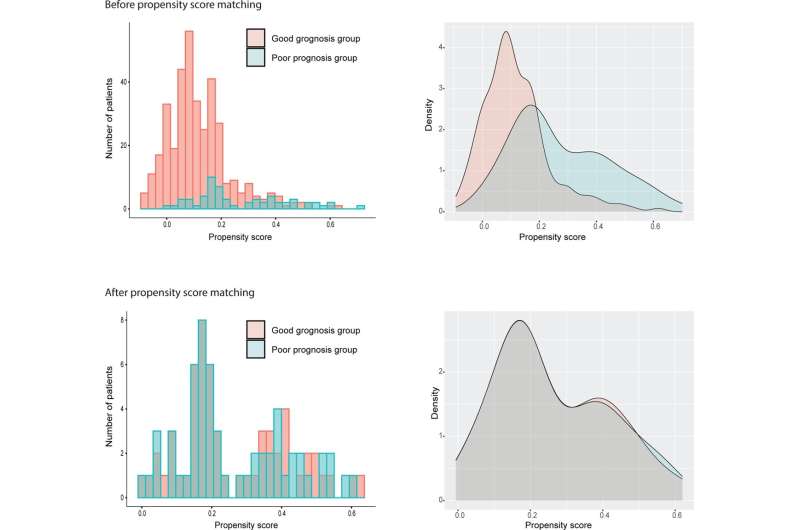
Corresponding author Michael H. Roehrl, MD, Ph.D., MBA, chair of the department of Pathology at BIDMC, and colleagues used archived tissue samples to sort cancer cases into good or poor prognosis groups based on patient survival information. The groups were then compared to uncover prognostic protein or transcript biomarkers.
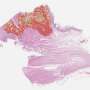
One dataset was derived from proteomic expression profiles of 544 surgically resected CRC tissue obtained from pathology archives of a large academic medical center. This cohort comprised 367 patients who survived more than three years without recurrence (good outcome group), and 60 patients who had recurrence within three years (poor outcome group). The second group consisted of RNA sequencing profiles of 163 CRC cases, 130 of whom were in the good prognosis group and 33 of whom were considered in the poor prognosis group.
Unlike conventional analysis which typically compares randomized cohorts of cancer and normal tissues, PSM enabled direct comparison between patient characteristics, uncovering new prognostic biomarkers. By creating optimally matched groups to minimize confounding effects, this study demonstrated that PSM enables robust extraction of significant biomarkers while requiring fewer cancer cases and smaller overall patient cohorts.
More information: Masaki Maekawa et al, Propensity score matching as an effective strategy for biomarker cohort design and omics data analysis, PLOS ONE (2024). DOI: 10.1371/journal.pone.0302109