This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Computational tool integrates transcriptomic data to enhance breast cancer diagnosis and treatment
Addressing problems with diagnosing and treating breast cancer, scientists at EPFL have developed EMBER, a tool that integrates breast cancer transcriptomic data from multiple databases. EMBER can improve precision oncology by accurately predicting molecular subtypes and therapy responses.
Breast cancer is the most frequently diagnosed cancer worldwide. However, it is not a uniform disease; it comes in different subtypes, which need to be accurately identified for doctors to effectively tailor treatments to individual patients.
Cancer subtyping has traditionally been performed with histological staining (immunohistochemistry), which visually identifies specific markers that can classify a tumor into a specific subtype.
But in recent years, another method has revolutionized the subtyping of breast cancer: high-throughput transcriptomic profiling, which looks instead at the gene activity of cancer cells by detecting the sum total of messenger RNAs in each cell (messenger RNA corresponds to the sequence of a gene, and read by a ribosome in the process of synthesizing a protein).
Transcriptomics rely on RNA sequencing ("RNAseq"), a booming molecular biology technology that quickly "reads" the sequence of the RNA string. "A lot of patient breast cancer samples have been subjected to global gene expression profiling by consortia, and there are actually three major public databases with thousands of patient samples explored by researchers worldwide," says EPFL Professor Cathrin Brisken.
She adds, "We have learnt a lot from various analyses and there are suggestions that RNA sequencing—as it is becoming cheaper—could be applied to routine clinical practice and help with diagnosis and decision making. However, this is hampered by the fact that RNAseq analysis typically required big batches of samples to be processed at the same time and samples from different platforms are difficult to compare."
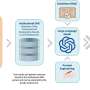
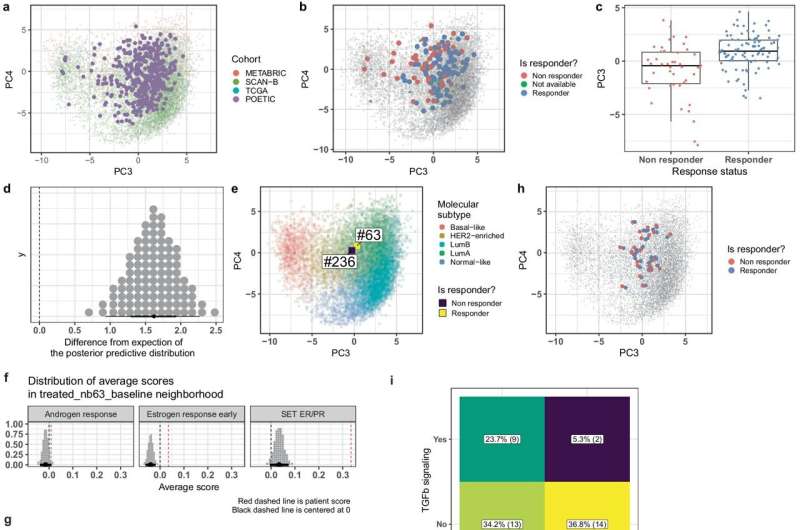
Now, under the umbrella of the 4.2 Mio EU transdisciplinary Ph.D. training network CANCERPREV Brisken coordinated EMBER ("molecular EMBeddER") was conceived. It is a computational tool that brings together over 11,000 breast cancer transcriptomes to predict cancer subtypes on a single-sample basis and accurately captures key biological pathways, offering superior predictive power for therapy responses.
EMBER was developed by Carlos Ronchi while studying for his Ph.D. at Brisken's lab. "Carlos developed an approach by which he places the major databases into a common space," says Brisken. "He showed that he can add additional cohorts into this space and, most excitingly, even individual samples. The position of a sample in this 'EMBER' space provides additional biological information."
To create EMBER, the researchers developed a statistical model that integrates both RNA-seq and microarray data from prominent datasets, including TCGA and METABRIC. The research is published in the journal npj Breast Cancer.
They focused on early-stage breast cancer patients, normalizing the data to bring it onto a common scale. By selecting the 1,000 most variable genes and using 44 stable genes for normalization, they preserved critical gene expression characteristics.
The team validated EMBER using independent patient cohorts and applied it to clinical trial data, such as the POETIC trial, where it identified potential mechanisms of therapy resistance, such as increased androgen receptor signaling and decreased TGFβ signaling. EMBER also effectively captured the five molecular subtypes of breast cancer and key biological pathways like estrogen receptor signaling and cell proliferation.
One significant discovery was that the EMBER-based estrogen receptor signaling score outperformed the immunohistochemistry-based ER index, which is currently used in clinical practice. This finding suggests that EMBER can more accurately predict responses to endocrine therapy.
By providing a unified space for breast cancer transcriptomic data, EMBER allows for a more nuanced understanding of molecular subtypes and therapy responses. This could lead to more personalized treatment plans and better outcomes for patients with ER+ breast cancer.
EMBER also offers a potential pathway for integrating RNA sequencing into standard diagnostic practices, paving the way for more comprehensive and cost-effective cancer diagnostics. This approach not only enhances precision oncology but also provides a robust framework for future research and clinical applications.
More information: Carlos Ronchi et al, EMBER creates a unified space for independent breast cancer transcriptomic datasets enabling precision oncology, npj Breast Cancer (2024). DOI: 10.1038/s41523-024-00665-z