This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Artificial intelligence method could advance gene mutation prediction in lung cancer
Research presented today suggests an artificial intelligence tool called DeepGEM may provide an advancement in genomic testing that offers an accurate, cost-effective, and timely method for gene mutation prediction from histopathology slides.
The research was presented today at the IASLC 2024 World Conference on Lung Cancer by Professor Wenhua Liang, from the China State Key Laboratory of Respiratory Disease and National Clinical Research Center for Respiratory Disease, the First Affiliated Hospital of Guangzhou Medical University, China.
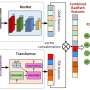
Accurate detection of driver gene mutations is essential for effective treatment planning and prognosis prediction in lung cancer. Traditional genomic testing, which relies on high-quality tissue samples, is often time-consuming and resource-intensive, limiting accessibility, particularly in low-resource settings. Addressing this gap, Prof. Liang and his team used DeepGEM, which uses routinely acquired histology slides to predict gene mutations, significantly enhancing accessibility and efficiency in mutation screening.
Prof. Liang and his colleagues analyzed datasets from 16 centers and 3,658 patients. This dataset, which includes paired pathological images and gene mutation data, was complemented by publicly available datasets from The Cancer Genome Atlas.
DeepGEM was initially trained and evaluated on an internal dataset of 1,717 patients and subsequently tested on an external dataset from 15 additional centers with 1,719 patients and a public dataset of 535 patients. To assess generalizability, the model was also tested on a lymph node metastases dataset consisting of 331 biopsies.
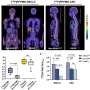
DeepGEM demonstrated performance with a median area under the curve of 0.938 for excisional biopsies and 0.891 for aspiration biopsies in the internal dataset. On the multi-center external dataset, the model achieved median AUCs of 0.859 for excisional biopsies and 0.826 for aspiration biopsies.
The model was further validated with an AUC of 0.874 on the TCGA dataset, showcasing its effectiveness across diverse racial backgrounds. Importantly, DeepGEM's ability to predict mutations from primary biopsies extended to lymph node metastases, indicating its potential for prognostic prediction of targeted therapies.
According to Prof. Liang, DeepGEM also provides interpretable results, generating spatial gene mutation maps at the single-cell level, which have been validated against immunohistochemistry results, underscoring the model's accuracy and reliability.
"Compared to previous studies, DeepGEM achieved robust and superior predictive performance across various genes, validating the largest multicenter datasets to date. The rapid prediction capabilities of DeepGEM allow for quicker decision-making in treatment, enabling patients with severe symptoms to receive targeted therapies sooner," Prof. Liang reported.
"Furthermore, it presents opportunities for multi-gene mutation detection and precision treatment in economically underdeveloped areas where genomic testing is unaffordable. This innovative approach has the potential to transform the clinical management of lung cancer patients, making advanced genomic insights more accessible and actionable."