When is a gene not a gene? New catalog helps identify gene variations associated with disease
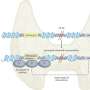
![Distribution of recessive disease genes (blue) and LoF-tolerant genes (red) compared to all protein-coding genes (gray) [DOI:10.1126/science.1215040] When is a gene not a gene? New catalog helps identify gene variations associated with disease](https://scx1.b-cdn.net/csz/news/800a/2012/whenisagenen.jpg)
A high-quality reference catalogue of the genetic changes that result in the deactivation of human genes has been developed by a team of researchers. This catalogue of loss-of-function (LoF) variants is needed to find new disease-causing mutations and will help us to better understand the normal function of human genes. In addition, the researchers report that each of us is carrying around 20 genes that have been completely inactivated.
The team refined previous estimations of possible LoF variants by excluding more than half. They accomplished this by identifying errors and real variants that did not seem to affect gene function and eliminating them from the list. They also developed a method of determining whether or not a newly-identified variant could be a likely cause of disease.
Loss of function variants are genetic changes that are predicted to severely disrupt the function of genes. They are known to cause severe human diseases such as muscular dystrophy and cystic fibrosis. Previous genome sequencing projects have suggested that hundreds of these variants are present in the DNA of even perfectly healthy individuals, but could not tell exactly how many.
"The key questions we focused on for this study were: how many of these LoF variants were real and how large a role might they play in human disease?" explains Dr Daniel MacArthur, first author from the Wellcome Trust Sanger Institute. "We looked at nearly 3000 putative LoF variants in the genomes of 185 people from Europe, East Asia and West Africa who were participants of the 1000 Genomes Project."
Working as part of the 1000 Genomes Project, the team developed a series of filters to identify common errors. The filters revealed that 56% of the 3000 LoFs were unlikely to seriously affect gene function. But of the true LoF variants, 100 are typically found in the genome of each European and 20 affect both copies of the gene, and are thus predicted to result in complete loss of gene function.
"We identified 253 genes that can be completely inactivated in one or more participant. This shows that at least 1% of human genes can be shut down without causing serious disease", explains Professor Mark Gerstein, co-author from Yale University. "We were able to use the differences between such "LoF-tolerant" genes and known human disease genes to develop a way of predicting whether or not a newly-discovered change in a gene is likely to be severely disease-causing."
The team found some of these LoFs are quite common and are unlikely to have a significant effect on health. For instance, some can affect the way in which we detect smells or how sensitive we are to sour taste. However, they found that the majority of the LoF variants are rare, with half of them being seen only once in the 185 people. This suggests that most of these variants can be quite harmful.
"Our research will be beneficial for current DNA sequencing studies underway in disease patients," says Dr Chris Tyler-Smith, lead author from the Wellcome Trust Sanger Institute. "In addition, we provide a list of over 1000 loss-of-function variants, and in most cases little or nothing is known about how these genes work or what they do. By studying the people carrying them in detail, we should get new insights into the function of many poorly-known human genes"
The team's long term goal is to study the potential effects all LoF variants have on humans. They will do this by looking at them in people with different diseases, as well as healthy people who have been measured for many different traits.
More information: McArthur et al 'A Systematic Survey of Loss-of-Function Variants in Human Protein-Coding Genes' Published in Science on 17 February 2012. dx.doi.org/10.1126%2Fscience.1215040