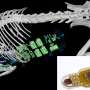
Translation error tracked in the brain of dementia patients
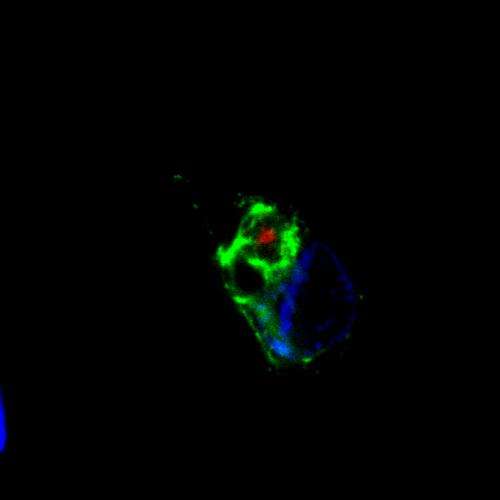
In certain dementias silent areas of the genetic code are translated into highly unusual proteins by mistake. An international team of scientists including researchers from the German Center for Neurodegenerative Diseases in Munich and the Ludwig-Maximilians-Universitat present this finding in the online edition of Science.
The proteins that have now been identified shouldn't actually exist. Nevertheless, they build the core of cellular aggregates whose identity has been enigmatic until now. These aggregates are typically associated with hereditary neurodegenerative diseases including variants of frontotemporal dementia (FTD), also known as frontotemporal lobar degeneration (FTLD), and amyotrophic lateral sclerosis (ALS). They are likely to be damaging and might be a target for therapy.
FTD and ALS are part of a group of neurodegenerative diseases that show a broad and overlapping variety of symptoms: Patients often suffer from dementia, personality changes and may also be affected by language abnormalities and movement disorders. The problems often arise before the age of 65 without a clear cause. However, about 30 percent of cases are linked to a genetic cause. In Europe approximately 10 percent of patients show a common genetic feature: In their DNA (the carrier of the genetic code) a particular short sequence appears in numerous copies one after another. Furthermore, proteins of unknown identity accumulate inside the brain of these patients. As it turns out both findings are directly related – that is what the team of researchers including molecular biologists Dieter Edbauer and Christian Haass has now been able to show.
"We have found that the proteins are linked to a genetic peculiarity which many patients have in common. At a certain location inside the gene C9orf72 there are several hundred repeats of the sequence GGGGCC, while healthy people display less than 20 such copies," explains Prof. Edbauer, who researches at the DZNE and the LMU. "But it is surprising that these proteins are actually made, because these repeats fall into a region of the DNA that should not be translated into proteins."
An area of DNA assumed to be silent
The DNA holds the blueprints for building proteins. In general, the beginning of such a blueprint is indicated by a certain molecular start signal, but the usual signal is missing in this case. The region of DNA comprising the numerous repeats should therefore not be translated into proteins. It seems that the process of protein synthesis is initiated in a non-textbook way. "Although quite rare there are two known alternatives to the common mechanism. Which procedure applies here, we don't know yet," says Prof. Haass, Site Speaker of the DZNE in Munich and chair of Metabolic Biochemistry at LMU.
Nevertheless, in cell culture experiments the researchers were able to show that long repeats of the sequence GGGGCC may in fact lead to the production of proteins, even though the usual start signal is missing. Furthermore, they identified the same proteins in the particles that typically accumulate in the brain of patients. The scientist could also identify their composition: They turned out to be dipeptid-repeat proteins, which comprise a very large number of identical building blocks.
"These are very extraordinary proteins that usually don't show-up in the organism," Edbauer notes. "As far as we know, they are completely useless and scarcely soluble. Therefore, they tend to aggregate and seem to damage the nerve cells. We haven't formally proven toxicity, but there is ample evidence." Because of their peculiarity these proteins might be an interesting target for new therapies. "As the mechanism of their production is so unusual, we may find ways to inhibit their synthesis without interfering with the formation of other proteins. One could also try to block their aggregation and accelerate their decomposition."
The scientists have applied for a patent and are pursuing a major goal. "At the DZNE in Munich it is our dream to develop a therapy against these devastating diseases," Haass and Edbauer conclude.
More information: The C9orf72 GGGGCC Repeat is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS, Kohji Mori et al., Science Express, doi: 10.1126/science.1232927