Researchers identify genetic variants predicting aggressive prostate cancers
Researchers at Moffitt Cancer Center and colleagues at Louisiana State University have developed a method for identifying aggressive prostate cancers that require immediate therapy. It relies on understanding the genetic interaction between single nucleotide polymorphisms (SNPs). The goal is to better predict a prostate cancer's aggressiveness to avoid unnecessary radical treatment.
Their study was published in the online journal PLOS ONE in April.
According to the authors, prostate cancer accounts for 20 percent of all cancers and 9 percent of cancer deaths. It is the most common cancer and was the second leading cause of cancer death in American men in 2012.
"For most prostate cancer patients, the disease progresses relatively slowly," said study co-author Hui-Yi Lin, Ph.D., assistant member of the Chemical Biology and Molecular Medicine Program at Moffitt. "However, some cases grow aggressively and metastasize. It is often difficult to tell the difference between the two."
The two treatment options for aggressive prostate cancer—radical surgery and radiation therapy—have negative side effects, such as incontinence and erectile dysfunction. It is why the authors believe there is an urgent need for biomarkers that can identify or predict aggressive types of prostate cancer.
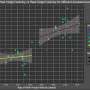
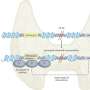
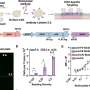
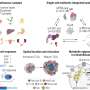
Through examining combinations of genetic variants, or SNP-SNP interactions, the researchers have identified and validated several genetic changes that are related to prostate cancer aggressiveness. Their work also shows that the epithelial growth factor receptor may be the hub for these interactions because it is involved in the growth of blood vessels (angiogenesis), which in turn stimulates tumor growth.
"Our findings identified five SNP-SNP interactions in the angiogenesis genes associated with prostate cancer aggressiveness," explained study co-author Jong Y. Park, Ph.D., associate member of Moffitt's Cancer Epidemiology Program. "We successfully detected the genotype combinations that put patients at risk of aggressive prostate cancer and then explored the underlying biological associations among angiogenesis genes associated with aggressive prostate cancer."
The researchers concluded that the gene network they constructed based on SNP-SNP interactions indicates there are novel relationships among critical genes involved in the angiogenesis pathway in prostate cancer.
"Our findings will help physicians identify patients with an aggressive type of prostate cancer and may lead to better personalized treatment in the future," Park said.
More information: www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0059688