Researchers explore new treatments for a leading genetic cause of infant deaths
Researchers at Iowa State University have identified an RNA structure in humans that could lead to a new treatment for spinal muscular atrophy, the leading genetic cause of death in babies and young children.
Ravindra Singh, a professor of biomedical sciences in the ISU College of Veterinary Medicine, is the lead author of a paper published this week in the journal Nucleic Acids Research that details the discovery of a novel therapeutic target that could be modified by medication, leading to new treatment possibilities for the disease.
Spinal muscular atrophy results from the loss or mutation of a gene called Survival Motor Neuron 1, often referred to as SMN1. If SMN1 is deleted or doesn't function properly, not enough SMN protein is produced, giving rise to the disease.
Luckily, the vast majority of humans have a nearly identical gene, referred to as SMN2, which can function as a substitute. But a critical portion of SMN2 is sometimes erroneously removed during the process known as splicing, or when pre-mRNA is turned into mRNA by getting rid of non-functioning parts of the gene.
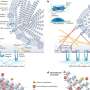
In the new paper, Singh and his colleagues have discovered an RNA structure exclusively formed by intronic sequences, or sequences that are removed during splicing. By targeting that structure, it may be possible to develop new treatments that prevent the mistake in the splicing process that causes the loss of function of SMN2, Singh said. If so, this is the first time a deep intronic structure can be targeted for therapy.
"About a quarter of a person's genome is made up of introns, or non-coding sequences, that must be removed through splicing throughout life," Singh said. "We've found an RNA structure that aberrantly promotes the escape of one of the coding sequences through splicing."
Singh cautioned that development of a new treatment would have to go through years of clinical trials and further study, but the bottom line is that the research could result in a new way to cure spinal muscular atrophy. In fact, private companies have shown interest in negotiating with Iowa State to begin development of a drug based on the research, Singh said.
He said the paper required about five years of painstaking work, testing hundreds of mutations to individual gene sequences one at a time to see if they have an effect on splicing.
"The process involves a lot of trial and error," he said.
Studying RNA structures within the non-coding portion of the human genome is still a relatively untouched frontier with much left to teach us, Singh said. It appears that RNA structures hold enormous information, and new techniques for studying RNA structures are unlocking new possibilities that could have major implications for how we treat genetic diseases, he said.
"In many ways this is still a very poorly understood field," Singh said. "But around half of all genetic diseases are a result of errors in splicing, so we have much to gain from answering these questions."
More information: nar.oxfordjournals.org/content … 7/14/nar.gkt609.full