Silent RNAs express themselves in ALS disease
RNA molecules, used by cells to make proteins, are generally thought to be "silent" when stowed in cytoplasmic granules. But a protein mutated in some ALS patients forms granules that permit translation of stored RNAs, according to a study in The Journal of Cell Biology. The finding identifies a new mechanism that could contribute to the pathology of the disease.
ALS, often referred to as "Lou Gehrig's Disease," is a progressive neurodegenerative disease that affects nerve cells in the brain and the spinal cord. Although the cause of ALS is not completely understood, researchers have been increasingly focused on RNA processing as an important cause of disease symptoms.
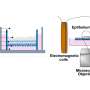
RNAs are gregarious, clustering with other RNA molecules and proteins to form RNP (ribonucleoprotein) complexes. RNPs then can gather into larger, more complex structures within the cell called granules. There are several kinds of granules, some that are always present and others that appear under stress, and researchers have generally thought that RNAs in granules are not translated into proteins.
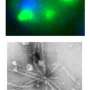
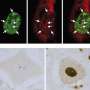
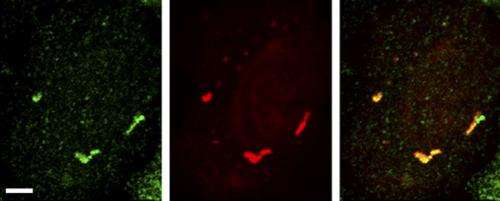
A team of researchers led by Stavroula Mili from the National Cancer Institute in Bethesda, Maryland, took a closer look at the functions of a protein called Fus, which is mutated in some ALS patients and causes large RNA granules to form in the patients' cells. The researchers demonstrated that Fus normally promotes the translation of RNA found in RNPs localized in cell protrusions. But abnormal versions of Fus found in ALS patients have broader effects. Cells engineered to produce mutant Fus protein harbor cytoplasmic granules that are similar to those found in the cells of ALS patients. The researchers anticipated that RNAs in the granules would be silent, but they instead discovered that the cells translated several of the RNAs into proteins.
The results suggest a new mechanism that could potentially drive ALS, in which misdirection of RNA translation, rather than RNA silencing, might contribute to disease.
More information: Yasuda, K., et al. 2013. J. Cell Biol. DOI: 10.1083/jcb.201306058