Scientists uncover molecule that keeps pathogens like salmonella in check
Scientists at UT Southwestern Medical Center have found a potential new way to stop the bacteria that cause gastroenteritis, tularemia and severe diarrhea from making people sick.
The researchers found that the molecule LED209 interferes with the biochemical signals that cause bacteria in our bodies to release toxins.
"What we have here is a completely novel approach to combating illness," said Dr. Vanessa Sperandio, associate professor of microbiology and biochemistry at UT Southwestern and senior author of a study available online today and in a future issue of Science.
Though many antimicrobial drugs are already available, new ones are needed to combat the increasing microbial resistance to antibiotics. In addition, treating some bacterial infections with conventional antibiotics can cause the release of more toxins and may worsen disease outcome.
Scientists have known for decades that millions of potentially harmful bacteria exist in the human body, awaiting a signal that it's time to release their toxins. Without those signals, the bacteria pass through the digestive tract without infecting cells. What hasn't been identified is how to prevent the release of those toxins, a process that involves activating virulence genes in the bacteria.
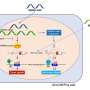
In the new study, UT Southwestern researchers describe how LED209 blocks the bacterial receptor for these signals. In 2006, the UT Southwestern researchers were the first to identify the receptor QseC sensor kinase, which is found in the membrane of a diarrhea-causing strain of Escherichia coli. This receptor receives signals from human flora and hormones in the intestine that cause the bacteria to initiate infection.
In studies in vitro, Dr. Sperandio and her colleagues found that LED209 blocked the QseC sensors in E coli, Salmonella and Francisella tularensis bacteria, preventing them from expressing virulence traits. Using mice models of infection, the researchers also showed that LED209 blocks pathogenesis of Salmonella and F tularensis, preventing them from causing disease in these animals.
Though the researchers limited the study to three pathogens, they believe drugs that target QseC could have a broader spectrum because the sensor exists in at least 25 important animal and plant pathogens including Erwinia, which causes plant rot; Legionella pneumophila, which causes Legionnaires' disease; and Haemophilus influenzae, which causes lung infections.
Unlike conventional antibiotics, which work by killing bacteria, LED209 allows the pathogen to grow but not become virulent and make the host sick. Dr. Sperandio said killing the bacteria or inhibiting their growth just "angers" some bacteria and causes them to release toxins.
"The sensors in bacteria are waiting for the right signal to initiate the expression of virulent genes," she said. "Using LED209, we blocked those sensing mechanisms and basically tricked the bacteria to not recognize that they were within the host. When we did that, the bacterial pathogens could not effectively cause disease in the treated animals."
Allowing the pathogen to survive also makes it less likely to develop resistance to medical treatments.
"What makes this current study unique is that we showed the drug working in three different pathogens," Dr. Sperandio said. "Prior studies generally focused on one."
In early 2008, UT Southwestern received a five-year, $6.5 million grant from the National Institute of Allergy and Infectious Diseases to develop a new antimicrobial compound to target bacterial pathogens such as Salmonella, E coli and F tularensis. Dr. Sperandio is the principal investigator.
"Only a few new antibiotics have reached the market in recent years," Dr. Sperandio said. "Because LED209 has never been used as an antibiotic, it's a completely different type of drug. In addition, its target, QseC, is also different from the current antimicrobial drug targets. This study demonstrates that LED209 has promise in fighting at least three pathogens and likely many more."
Identifying LED209 was accomplished by using a high throughput screen of 150,000 compounds in UT Southwestern's Small Molecular Library. The screening process was set up to find molecules that wouldn't activate the virulence genes in a strain of E coli known as enterohemorrhagic E coli 0157:H7, or EHEC. Additional rounds of screening resulted in a pool of 75 potential inhibitors, from which LED209 was selected partly because of its potency.
The team's next step is to understand further LED209's structure and how it functions. The researchers plan to modify the drug to develop customized formulations.
"What we have right now works really well for systemic infections and it's very potent, but we also need non-absorbable molecules to treat noninvasive pathogens such as EHEC, which stays in the intestine," Dr. Sperandio said.
Source: UT Southwestern Medical Center