New technique comprehensively generates three-dimensional maps of gene expression in the brain
A research team led by Yuko Okamura-Oho and Hideo Yokota of the RIKEN Advanced Science Institute, Wako, has developed a novel technique for three-dimensional (3D) mapping of gene expression patterns onto brain structures. The technique, known as transcriptome tomography, combines tissue sectioning with microarray technology and produces comprehensive maps of the density and location of gene expression, which have a higher resolution than the maps produced by existing methods.
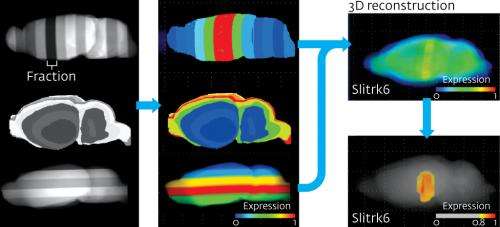
To produce their first dataset, the researchers sliced six mouse brains into five micrometer sections, in each of three anatomical planes. They collected the sections in batches of 200 to produce 'fractions' of 1-millimeter thickness that were used for microarray analysis. They then treated 61 such fractions with more than 36,000 RNA probes and reconstructed the data to produce 3D maps of gene expression throughout the whole mouse brain.
Transcriptome tomography is semi-automated, making it more cost-effective and faster than existing manual approaches—it took the researchers just one month to generate the first dataset. The technique can also be used to map the tissue distribution of any type of biological molecule, such as proteins, lipids, sugars and microRNAs.
Okamura-Oho and her colleagues validated the technique by comparing their first dataset to pre-existing ones generated by other methods. They also analyzed the expression patterns of the Huntingtin gene, which when mutated causes Huntington's disease, a progressive neurodegenerative condition characterized by the death of neurons in the basal ganglia, followed by cell death in the cerebral cortex.
The analysis revealed that Huntingtin was expressed at high levels in brain regions known to be severely affected by the condition, such as the basal ganglia, but at significantly lower levels in areas that are less vulnerable, such as the midbrain and cerebellum. "We could make expression maps in 20-times higher resolution comparable to MRI. Such maps have the potential to reveal more detailed disease-related abnormalities with continuous technical advancing," says Okamura-Oho.
Transcriptome tomography datasets can be uploaded to Waxholm Space, a co-ordinate-based space for open resources. The space facilitates the creation of researchers' own datasets that can then be shared and analyzed in the space.
More information: Okamura-Oho, Y., Shimokawa, K., Takemoto, S., Hirakiyama, A., Nakamura, S., Tsujimura, Y., Nishimura, M., Kasukawa, T., Masumoto, K., Nikaido, I. et al. Transcriptome tomography for brain analysis in the web-accessible anatomical space. PLoS ONE 7, e45373 (2012). www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0045373